Gene
KWMTBOMO03265
Pre Gene Modal
BGIBMGA013608
Annotation
PREDICTED:_activin_receptor_type-2B-like_[Bombyx_mori]
Full name
Serine/threonine-protein kinase receptor
+ More
Receptor protein serine/threonine kinase
Receptor protein serine/threonine kinase
Location in the cell
Extracellular Reliability : 1.26
Sequence
CDS
ATGAAGTTCCTACACATAATCTTGCTTGTGCTTGGTATGCATGCAATAACACTGGCGGCTCCACCATCACAAGAAAACCTCCCACCGAGGCCGAAAGAAGGCGGAGTAACCACCAGATACTGCGAATTCTACAATAGCTCCACTCCTAGCTGTAATCCAGCAACGAACTCTTCATGCGTACCTGAAACAATTATAGAGGAATGCCATCCATTCGAAGCGGAAAAAAGTGTACACTGCTTTGTGTTGTGGCAGTTTGATAACGAAATGTACACCGCATATCCTAAAGTCAAAGGATGTTTCGTCGATACAGTCTCGTGTAAAGACAGAGCATCCAACTGCACCCTCCACAATACGCTACTGCCACTTCTACACTGCTGTTGCGAAGGAGACATGTGCAACAAAAATCTTACGTTCCCCAATCCGAAATCATTTATGACACAAACAGATAATCCCAAAGAACACAGTACTGTCGCCTCCCCAATTCCCGCTGCAATGCCGGACGTCATTGCGTACACGCTGACACCGTTGTTCCTGCTATTCGCTGTGTTAGTCGGATGCTACCTGTTATACAGGAAACGTAAAGGCAGCTCTTTTGCTGAACTAGCCAGCGGCGAGAATTCTCTGTCCAGGCCTCCGTCGGCGGGCGGCGGTATGGAAAATGAATCCGGCGTGTTAGTCACTCAGGTGGCGCTCTGTGAAGTGAGAGCCCGCGGGAGATTTGGAGCGGTTTGGCGAGCGAGACTAGGACAAAAGGACGTGGCGGTTAAAGTGTTTCCCCTACAGGACAAGCAGTCGTGGTTAGCCGAGCAAGAGGTGTTCCGACTGCCCAGGATGGATCATCCCGACATTTTGCAGTACATCGGCGTTGATAAAACTGGCGATAATCTACAGGCAGAATATTGGCTCATCACAGCTTATCATGAGAAGGGCTCCCTATGCGACTACCTGAAAGCTAACACGCTAACTTGGGCTAAAGCGTGGCGTATAGCAGCTTGCGTGGCTCGGGGTCTGTCCCACCTGCACGAAGAAGGCGGTGGCAAACCCGCAATAGCGCACAGGGACTTTAAATCCAAAAACGTACTGTTGAAAGCGGATTTGAGCGCGTGCATCGCTGATTTCGGCCTAGCTCTGATATTCGTGCCGGCAAGAGGCTGTGGGGATGCTCACGGCCAGGTCGGAACAAGACGGTACATGGCTCCGGAGGTCCTGGACGGAGCTATCAACTTCACTAAAGACGCCTTCTTGAGGATTGACATGTACGCGTGCGCGCTGGTGCTATGGGAAATAGCTTCGAGATGCAGTGACGTAGGCGCAGACGATACGTCCCAGTACCGTTTGCCGTTGGAAGAAGAAGTCGGATCTCATCCGAGCCTCGAGGAGATGCAGGAAGCGGTCGTGCAAAGGAAACTCCGTCCGCACATACCGCAGCACTGGAGAGACCACCCGGGCATGTCAGTGCTGTGCGACACCATGGAGGAGTGCTGGGACCACGACGCGGAGGCGCGGCTGTCTGCGTCGTGCGTGCTGGAGCGCGTGGCCGCCCAGCGCGCCCCGTGCTCGGACAGTGCGCCGCTGCTCCTCCACCCCCCGCCCTGCTGA
Protein
MKFLHIILLVLGMHAITLAAPPSQENLPPRPKEGGVTTRYCEFYNSSTPSCNPATNSSCVPETIIEECHPFEAEKSVHCFVLWQFDNEMYTAYPKVKGCFVDTVSCKDRASNCTLHNTLLPLLHCCCEGDMCNKNLTFPNPKSFMTQTDNPKEHSTVASPIPAAMPDVIAYTLTPLFLLFAVLVGCYLLYRKRKGSSFAELASGENSLSRPPSAGGGMENESGVLVTQVALCEVRARGRFGAVWRARLGQKDVAVKVFPLQDKQSWLAEQEVFRLPRMDHPDILQYIGVDKTGDNLQAEYWLITAYHEKGSLCDYLKANTLTWAKAWRIAACVARGLSHLHEEGGGKPAIAHRDFKSKNVLLKADLSACIADFGLALIFVPARGCGDAHGQVGTRRYMAPEVLDGAINFTKDAFLRIDMYACALVLWEIASRCSDVGADDTSQYRLPLEEEVGSHPSLEEMQEAVVQRKLRPHIPQHWRDHPGMSVLCDTMEECWDHDAEARLSASCVLERVAAQRAPCSDSAPLLLHPPPC
Summary
Catalytic Activity
[receptor-protein]-L-serine + ATP = [receptor-protein]-O-phospho-L-serine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
Similarity
Belongs to the protein kinase superfamily. TKL Ser/Thr protein kinase family. TGFB receptor subfamily.
Feature
chain Serine/threonine-protein kinase receptor
Uniprot
H9JVP5
A0A2A4JKH1
A0A2H1VND3
A0A2W1BGW3
A0A212EM83
A0A0N0PCA0
+ More
A0A194Q296 A0A0L7KQP8 A0A232F241 K7J138 A0A088ANA1 A0A067RJQ7 A0A2A3EBL9 A0A1B6FDB3 A0A151WES3 E2BIN9 F4WR66 A0A195BBP4 A0A151IQG0 A0A0L7RG85 A0A1B6CZD7 A0A3L8DEJ0 A0A1Y1ME07 A0A026WSB3 A0A1W4W8K5 A0A151J326 D6X465 A0A1B6M5B9 T1HXB4 A0A023F3N5 U4UJ22 A0A0N8CKI7 A0A0P6JKQ6 A0A158NUB9 A0A195FKB9 A0A0A9YX77 A0A154PCP5 A0A2H8TI06 A0A2L2YPG4 A0A3Q0IY43 A0A385XRK0 A0A087T9S8 U5EVS8 J9JVN2 A0A1Q3F5D7 A0A023ET35 A0A182N1J7 A0A1Z5KX47 T1IHR6 A0A182FIE5 W5JT00 A0A293M8C5 A0A2M4AC59 A0A2M4BJJ1 A0A182XXR6 A0A2M4BKG1 A0A1L8DJS6 A0A182I9J1 A0A182QZB7 A0A1L8DK39 A0A2R7WNY7 Q0IEL3 A0A084VIW5 A0A1S4FGD1 A0A1S4FMY1 Q172G8 A0A182K107 A0A336MK22 A0A336LB80 A0A182XEL0 B4PSR2 A0A182V0B1 Q7QE97 B4NK20 A0A182LIN0 B3P3L7 A0A0B4KG59 Q24468 Q24229 A0A182RH81 A0A182M3H7 A0A0P4W8U5 B4HEL0 B4QZT3 B4G5Q5 Q299T8 A0A182IST1 C5WLU3 A0A3B0JD99 A0A182W474 A0A1W4V5R5 A0A147BTK8 A0A1I8MWT9 B4JXY6 B3LW28 B4K9K1 A0A2S2Q7T7 A0A0K8VH74 A0A034VX52 B4LVW1
A0A194Q296 A0A0L7KQP8 A0A232F241 K7J138 A0A088ANA1 A0A067RJQ7 A0A2A3EBL9 A0A1B6FDB3 A0A151WES3 E2BIN9 F4WR66 A0A195BBP4 A0A151IQG0 A0A0L7RG85 A0A1B6CZD7 A0A3L8DEJ0 A0A1Y1ME07 A0A026WSB3 A0A1W4W8K5 A0A151J326 D6X465 A0A1B6M5B9 T1HXB4 A0A023F3N5 U4UJ22 A0A0N8CKI7 A0A0P6JKQ6 A0A158NUB9 A0A195FKB9 A0A0A9YX77 A0A154PCP5 A0A2H8TI06 A0A2L2YPG4 A0A3Q0IY43 A0A385XRK0 A0A087T9S8 U5EVS8 J9JVN2 A0A1Q3F5D7 A0A023ET35 A0A182N1J7 A0A1Z5KX47 T1IHR6 A0A182FIE5 W5JT00 A0A293M8C5 A0A2M4AC59 A0A2M4BJJ1 A0A182XXR6 A0A2M4BKG1 A0A1L8DJS6 A0A182I9J1 A0A182QZB7 A0A1L8DK39 A0A2R7WNY7 Q0IEL3 A0A084VIW5 A0A1S4FGD1 A0A1S4FMY1 Q172G8 A0A182K107 A0A336MK22 A0A336LB80 A0A182XEL0 B4PSR2 A0A182V0B1 Q7QE97 B4NK20 A0A182LIN0 B3P3L7 A0A0B4KG59 Q24468 Q24229 A0A182RH81 A0A182M3H7 A0A0P4W8U5 B4HEL0 B4QZT3 B4G5Q5 Q299T8 A0A182IST1 C5WLU3 A0A3B0JD99 A0A182W474 A0A1W4V5R5 A0A147BTK8 A0A1I8MWT9 B4JXY6 B3LW28 B4K9K1 A0A2S2Q7T7 A0A0K8VH74 A0A034VX52 B4LVW1
EC Number
2.7.11.30
Pubmed
19121390
28756777
22118469
26354079
26227816
28648823
+ More
20075255 24845553 20798317 21719571 30249741 28004739 24508170 18362917 19820115 25474469 23537049 21347285 25401762 26823975 26561354 24945155 28528879 20920257 23761445 25244985 17510324 24438588 17994087 17550304 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7697719 26109357 26109356 8415726 15632085 29652888 25315136 25348373
20075255 24845553 20798317 21719571 30249741 28004739 24508170 18362917 19820115 25474469 23537049 21347285 25401762 26823975 26561354 24945155 28528879 20920257 23761445 25244985 17510324 24438588 17994087 17550304 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7697719 26109357 26109356 8415726 15632085 29652888 25315136 25348373
EMBL
BABH01032296
BABH01032297
NWSH01001169
PCG72276.1
ODYU01003477
SOQ42323.1
+ More
KZ150071 PZC74058.1 AGBW02013858 OWR42610.1 KQ460650 KPJ13124.1 KQ459579 KPI99458.1 JTDY01007140 KOB65435.1 NNAY01001172 OXU24886.1 KK852470 KDR23233.1 KZ288292 PBC29173.1 GECZ01021577 JAS48192.1 KQ983238 KYQ46368.1 GL448520 EFN84433.1 GL888284 EGI63293.1 KQ976532 KYM81630.1 KQ976780 KYN08388.1 KQ414596 KOC69977.1 GEDC01018507 JAS18791.1 QOIP01000009 RLU18741.1 GEZM01036296 JAV82820.1 KK107119 EZA58551.1 KQ980298 KYN16824.1 KQ971410 EEZ97734.1 GEBQ01008879 JAT31098.1 ACPB03011000 GBBI01002717 JAC15995.1 KB632345 ERL93082.1 GDIP01122751 LRGB01000007 JAL80963.1 KZS21938.1 GDIQ01016002 JAN78735.1 ADTU01026290 ADTU01026291 KQ981512 KYN40848.1 GBHO01009459 GDHC01020140 GDHC01004116 JAG34145.1 JAP98488.1 JAQ14513.1 KQ434864 KZC08970.1 GFXV01001874 MBW13679.1 IAAA01042312 LAA09984.1 MG545146 AYC12380.1 KK114189 KFM61867.1 GANO01003274 JAB56597.1 ABLF02033113 GFDL01012306 JAV22739.1 GAPW01001150 JAC12448.1 GFJQ02007432 JAV99537.1 JH429991 ADMH02000457 ETN66398.1 GFWV01010476 MAA35205.1 GGFK01005046 MBW38367.1 GGFJ01004061 MBW53202.1 GGFJ01004137 MBW53278.1 GFDF01007371 JAV06713.1 APCN01000984 AXCN02002441 GFDF01007370 JAV06714.1 KK855179 PTY21304.1 CH477600 EAT38449.1 ATLV01013412 KE524855 KFB37909.1 CH477437 EAT40929.1 UFQT01000674 SSX26408.1 UFQS01002415 UFQT01002415 SSX13983.1 SSX33402.1 CM000160 EDW97558.2 AAAB01008847 EAA06869.4 CH964272 EDW85062.1 CH954181 EDV48905.1 AE014297 AGB95956.1 L38495 AAC41566.1 AAF55079.1 L22176 AAA03579.1 AXCM01000900 GDRN01077355 GDRN01077354 JAI62754.1 CH480815 EDW42167.1 CM000364 EDX12931.1 CH479179 EDW24921.1 CM000070 EAL27613.2 BT088908 ACT09405.1 OUUW01000005 SPP80354.1 GEGO01001316 JAR94088.1 CH916377 EDV90548.1 CH902617 EDV41561.1 CH933806 EDW16661.2 GGMS01004417 MBY73620.1 GDHF01020987 GDHF01014404 JAI31327.1 JAI37910.1 GAKP01012839 JAC46113.1 CH940650 EDW67566.1
KZ150071 PZC74058.1 AGBW02013858 OWR42610.1 KQ460650 KPJ13124.1 KQ459579 KPI99458.1 JTDY01007140 KOB65435.1 NNAY01001172 OXU24886.1 KK852470 KDR23233.1 KZ288292 PBC29173.1 GECZ01021577 JAS48192.1 KQ983238 KYQ46368.1 GL448520 EFN84433.1 GL888284 EGI63293.1 KQ976532 KYM81630.1 KQ976780 KYN08388.1 KQ414596 KOC69977.1 GEDC01018507 JAS18791.1 QOIP01000009 RLU18741.1 GEZM01036296 JAV82820.1 KK107119 EZA58551.1 KQ980298 KYN16824.1 KQ971410 EEZ97734.1 GEBQ01008879 JAT31098.1 ACPB03011000 GBBI01002717 JAC15995.1 KB632345 ERL93082.1 GDIP01122751 LRGB01000007 JAL80963.1 KZS21938.1 GDIQ01016002 JAN78735.1 ADTU01026290 ADTU01026291 KQ981512 KYN40848.1 GBHO01009459 GDHC01020140 GDHC01004116 JAG34145.1 JAP98488.1 JAQ14513.1 KQ434864 KZC08970.1 GFXV01001874 MBW13679.1 IAAA01042312 LAA09984.1 MG545146 AYC12380.1 KK114189 KFM61867.1 GANO01003274 JAB56597.1 ABLF02033113 GFDL01012306 JAV22739.1 GAPW01001150 JAC12448.1 GFJQ02007432 JAV99537.1 JH429991 ADMH02000457 ETN66398.1 GFWV01010476 MAA35205.1 GGFK01005046 MBW38367.1 GGFJ01004061 MBW53202.1 GGFJ01004137 MBW53278.1 GFDF01007371 JAV06713.1 APCN01000984 AXCN02002441 GFDF01007370 JAV06714.1 KK855179 PTY21304.1 CH477600 EAT38449.1 ATLV01013412 KE524855 KFB37909.1 CH477437 EAT40929.1 UFQT01000674 SSX26408.1 UFQS01002415 UFQT01002415 SSX13983.1 SSX33402.1 CM000160 EDW97558.2 AAAB01008847 EAA06869.4 CH964272 EDW85062.1 CH954181 EDV48905.1 AE014297 AGB95956.1 L38495 AAC41566.1 AAF55079.1 L22176 AAA03579.1 AXCM01000900 GDRN01077355 GDRN01077354 JAI62754.1 CH480815 EDW42167.1 CM000364 EDX12931.1 CH479179 EDW24921.1 CM000070 EAL27613.2 BT088908 ACT09405.1 OUUW01000005 SPP80354.1 GEGO01001316 JAR94088.1 CH916377 EDV90548.1 CH902617 EDV41561.1 CH933806 EDW16661.2 GGMS01004417 MBY73620.1 GDHF01020987 GDHF01014404 JAI31327.1 JAI37910.1 GAKP01012839 JAC46113.1 CH940650 EDW67566.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000215335 UP000002358 UP000005203 UP000027135 UP000242457 UP000075809 UP000008237 UP000007755 UP000078540 UP000078542 UP000053825 UP000279307 UP000053097 UP000192223 UP000078492 UP000007266 UP000015103 UP000030742 UP000076858 UP000005205 UP000078541 UP000076502 UP000079169 UP000054359 UP000007819 UP000075884 UP000069272 UP000000673 UP000076408 UP000075840 UP000075886 UP000008820 UP000030765 UP000075881 UP000076407 UP000002282 UP000075903 UP000007062 UP000007798 UP000075882 UP000008711 UP000000803 UP000075900 UP000075883 UP000001292 UP000000304 UP000008744 UP000001819 UP000075880 UP000268350 UP000075920 UP000192221 UP000095301 UP000001070 UP000007801 UP000009192 UP000008792
UP000215335 UP000002358 UP000005203 UP000027135 UP000242457 UP000075809 UP000008237 UP000007755 UP000078540 UP000078542 UP000053825 UP000279307 UP000053097 UP000192223 UP000078492 UP000007266 UP000015103 UP000030742 UP000076858 UP000005205 UP000078541 UP000076502 UP000079169 UP000054359 UP000007819 UP000075884 UP000069272 UP000000673 UP000076408 UP000075840 UP000075886 UP000008820 UP000030765 UP000075881 UP000076407 UP000002282 UP000075903 UP000007062 UP000007798 UP000075882 UP000008711 UP000000803 UP000075900 UP000075883 UP000001292 UP000000304 UP000008744 UP000001819 UP000075880 UP000268350 UP000075920 UP000192221 UP000095301 UP000001070 UP000007801 UP000009192 UP000008792
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JVP5
A0A2A4JKH1
A0A2H1VND3
A0A2W1BGW3
A0A212EM83
A0A0N0PCA0
+ More
A0A194Q296 A0A0L7KQP8 A0A232F241 K7J138 A0A088ANA1 A0A067RJQ7 A0A2A3EBL9 A0A1B6FDB3 A0A151WES3 E2BIN9 F4WR66 A0A195BBP4 A0A151IQG0 A0A0L7RG85 A0A1B6CZD7 A0A3L8DEJ0 A0A1Y1ME07 A0A026WSB3 A0A1W4W8K5 A0A151J326 D6X465 A0A1B6M5B9 T1HXB4 A0A023F3N5 U4UJ22 A0A0N8CKI7 A0A0P6JKQ6 A0A158NUB9 A0A195FKB9 A0A0A9YX77 A0A154PCP5 A0A2H8TI06 A0A2L2YPG4 A0A3Q0IY43 A0A385XRK0 A0A087T9S8 U5EVS8 J9JVN2 A0A1Q3F5D7 A0A023ET35 A0A182N1J7 A0A1Z5KX47 T1IHR6 A0A182FIE5 W5JT00 A0A293M8C5 A0A2M4AC59 A0A2M4BJJ1 A0A182XXR6 A0A2M4BKG1 A0A1L8DJS6 A0A182I9J1 A0A182QZB7 A0A1L8DK39 A0A2R7WNY7 Q0IEL3 A0A084VIW5 A0A1S4FGD1 A0A1S4FMY1 Q172G8 A0A182K107 A0A336MK22 A0A336LB80 A0A182XEL0 B4PSR2 A0A182V0B1 Q7QE97 B4NK20 A0A182LIN0 B3P3L7 A0A0B4KG59 Q24468 Q24229 A0A182RH81 A0A182M3H7 A0A0P4W8U5 B4HEL0 B4QZT3 B4G5Q5 Q299T8 A0A182IST1 C5WLU3 A0A3B0JD99 A0A182W474 A0A1W4V5R5 A0A147BTK8 A0A1I8MWT9 B4JXY6 B3LW28 B4K9K1 A0A2S2Q7T7 A0A0K8VH74 A0A034VX52 B4LVW1
A0A194Q296 A0A0L7KQP8 A0A232F241 K7J138 A0A088ANA1 A0A067RJQ7 A0A2A3EBL9 A0A1B6FDB3 A0A151WES3 E2BIN9 F4WR66 A0A195BBP4 A0A151IQG0 A0A0L7RG85 A0A1B6CZD7 A0A3L8DEJ0 A0A1Y1ME07 A0A026WSB3 A0A1W4W8K5 A0A151J326 D6X465 A0A1B6M5B9 T1HXB4 A0A023F3N5 U4UJ22 A0A0N8CKI7 A0A0P6JKQ6 A0A158NUB9 A0A195FKB9 A0A0A9YX77 A0A154PCP5 A0A2H8TI06 A0A2L2YPG4 A0A3Q0IY43 A0A385XRK0 A0A087T9S8 U5EVS8 J9JVN2 A0A1Q3F5D7 A0A023ET35 A0A182N1J7 A0A1Z5KX47 T1IHR6 A0A182FIE5 W5JT00 A0A293M8C5 A0A2M4AC59 A0A2M4BJJ1 A0A182XXR6 A0A2M4BKG1 A0A1L8DJS6 A0A182I9J1 A0A182QZB7 A0A1L8DK39 A0A2R7WNY7 Q0IEL3 A0A084VIW5 A0A1S4FGD1 A0A1S4FMY1 Q172G8 A0A182K107 A0A336MK22 A0A336LB80 A0A182XEL0 B4PSR2 A0A182V0B1 Q7QE97 B4NK20 A0A182LIN0 B3P3L7 A0A0B4KG59 Q24468 Q24229 A0A182RH81 A0A182M3H7 A0A0P4W8U5 B4HEL0 B4QZT3 B4G5Q5 Q299T8 A0A182IST1 C5WLU3 A0A3B0JD99 A0A182W474 A0A1W4V5R5 A0A147BTK8 A0A1I8MWT9 B4JXY6 B3LW28 B4K9K1 A0A2S2Q7T7 A0A0K8VH74 A0A034VX52 B4LVW1
PDB
2QLU
E-value=1.03489e-107,
Score=998
Ontologies
KEGG
GO
GO:0005524
GO:0043235
GO:0004675
GO:0046872
GO:0007179
GO:0016021
GO:0004674
GO:0046332
GO:0019838
GO:0005887
GO:0007389
GO:0005026
GO:0048179
GO:0032924
GO:0048185
GO:0034713
GO:0005024
GO:0007476
GO:0030718
GO:0007411
GO:0042078
GO:0030509
GO:0007391
GO:0007428
GO:0008258
GO:0009953
GO:0070724
GO:0007443
GO:0036122
GO:0017002
GO:0005886
GO:0001745
GO:0045705
GO:0006468
GO:0048666
GO:0007424
GO:0004672
GO:0048813
GO:0004702
GO:0007178
GO:0016020
GO:0005634
GO:0006270
GO:0042555
GO:0048384
GO:0038023
PANTHER
Topology
Subcellular location
Membrane
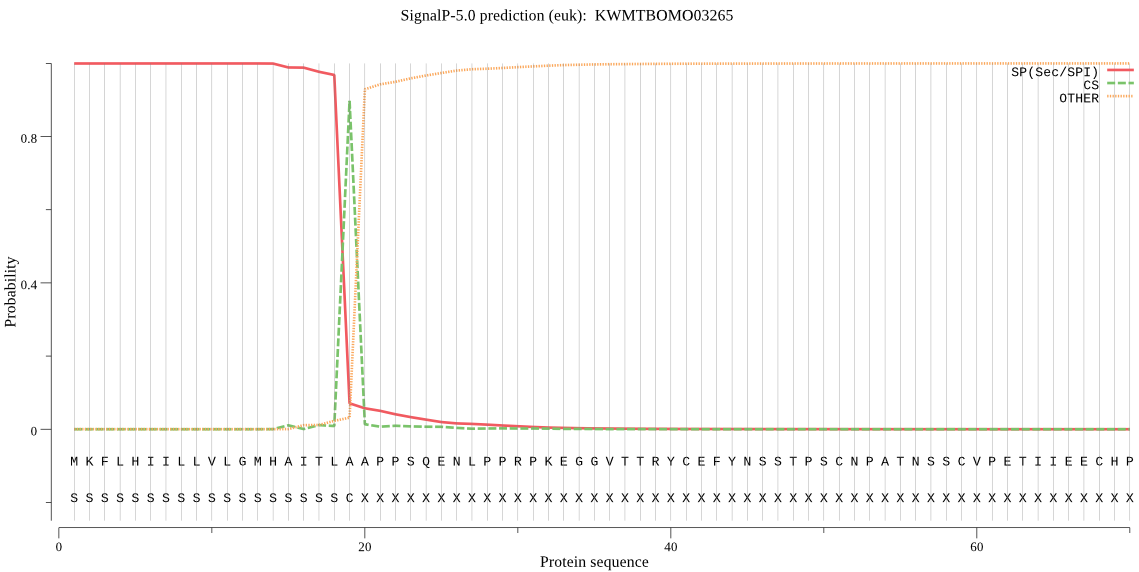
SignalP
Position: 1 - 19,
Likelihood: 0.999678
Length:
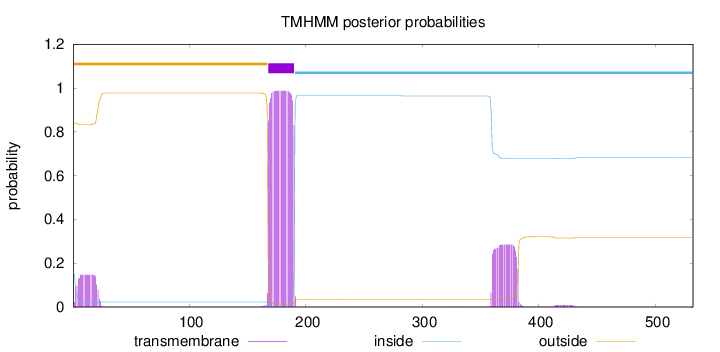
532
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
31.61468
Exp number, first 60 AAs:
2.76829
Total prob of N-in:
0.16069
outside
1 - 167
TMhelix
168 - 190
inside
191 - 532
Population Genetic Test Statistics
Pi
207.464052
Theta
187.735971
Tajima's D
0.338648
CLR
181.138187
CSRT
0.455027248637568
Interpretation
Uncertain
