Gene
KWMTBOMO03261
Pre Gene Modal
BGIBMGA013603
Annotation
PREDICTED:_protein_SMG8_[Bombyx_mori]
Full name
Protein SMG8
Alternative Name
Protein smg-8 homolog
Location in the cell
Nuclear Reliability : 1.324 PlasmaMembrane Reliability : 1.476
Sequence
CDS
ATGAAGGTTTTTAAATTAAATGATATACCAGAGTTTTCCAAAAGGGAGAGAATAGTAGTTGTCGGAATTATTGGCAAATCTCCTCACCGGTATCCAAACAAGACAACTCCACTACTTTCTGCAGTACAATCTCAAGAAATTGGCATCGAATGTCACTGGGATGAGCGCCGTGGTGTTCTGTTTCTCCATGCCCTCACGTATCTTGACACAAAACATCTTTCAGCCTTAGCTGCTGACTTGAATGAGAATTCCAAAAGCACAATTAAAGATGCAGATGCCTCAAATTGGCTTGTTGCATCGGGTGAACTAGCTATAGACTCATGCAAGATTATTGGATTGCTTTTTAATTTGTGTCATATTGTGGTGCTGTCATCACCCACCACTGTTTTTGATCTCGGATACCTACAATTATTTAAAGCTATCGATGCTTTTAGATTGGAAATAACAAACCGTACCTCAGTGGCCTTATCTGAAGCAGAGTTAGGTGGCACATGGATCTCGCATGGACGTTGTTGTTGCCCACGGCTACTGTTCCACATGCGTCGAGCTCCCGTACCTTTGAAACGTAATCCTGGAGCTTTAAAACGACTCGAACACTCCATTGAAGACCAACTTTATTCTATACTTAGAAAGTCTAGGATCATAACTAATGTTTGTGCAAAAGCCCTATTTGCAATACCGAAAAATGAAGAGTTTGTGTACATAAGCACTGATGAGGAAGTGTGTAACGCTCGCGACGTTAGCGTGCTGGTGAGCGGGCTGGTGCGCATGTGTGGCGGCGCGCAGCCCGACCGCCCCGCCGGGGAGAGGCCCGGCTTCGCGCAGTTCCTGCAGGGGCACATAGACTTGGCGTTCGCCGAAGGATTCGACGACAACGTCGGGAAATACGCGATGAGTACGTCATTCTTTGAGCTGCCGAGCGCAGAATCATGGCGTAAAGCAGCCGAGGTCCTGACGCCGCTGTACCTGGAGGATCCCGTGCAGGTGGACGAGGGCTCGGAGATCGGCCGCACTCTGTTCCTGATACTCGCCACTGACGTACGGTTCTCCCAGGCTCGATGCGCCAAGATATTACCAATAGCCCAGGCATCCTACACGGAGGGCATGCCGGCGCACTACTCCAGCCAGCACCACGCGCATAAGGTGAAGGTAGCCCTAGGAGTGCTGGAGACGATGGCCCGCGGTCCGCTGGCGGGCGGGGCTCGCGCGAGGCTGCAGGCCGCGTGTGACGTCACGTGGCGCTCCCGCAGTCTGTGCGAGGCGTTGTCCCTCACCGGACATCCGTGTATAGAACCCGAACACGACGGCGGCACGGAGCACTCGTCGGGCGTGCGCTACGTGTCGGCGTGCGCGTGCGGGCGCAGCAAGTGCTCGCGCGACGACCCGTACACGCCGCGCGCCGCCAACCTCGCCTTCTACTCGCTCGCGGCCGACCAGTGCGCGCACTGCCGCACGCTGCAGGCCGTCGCCTTCCCGGTCTTCCAGCCCTCCACGCCCACCTACAAAGCGGCATCAGTAAAAGGAGCACAAAGTCAAGAGGCCTACGAAAGTTCGACGGTCCGCGAGACGGAGGGGAACTCCGCGTCGGGGGGCGAGGGGGGAGAGGGGGACGAGTCGGGGGGCTGGTCCGCGCCGCACGCGCTGTCCCCGGGGTCGGGCGGGGACGACGACGAGGAGAACAAGTCCCCCGCAGCCGAGAACAGATACGTCGTGCCCGTTACTGGGGACTCTAATACTGATAAGATGATCCGCCGCCAGCCGTCGACGACGGAGTACCTGCCGGGCATGCTGCACACCGCCAGCCCGGCCGGCCTGCTGCCCGCCTACTGCTCCTGGAGCCTGGTGTGCGTGGGCGCCTCCTCTCTGTACTCGCACAGCATGGGCCTGCCGGACCACCTGCAGCCCGGCTTCCTGCCCCACACCAACTATCTGCTGCCCTGGGACTGCGCCGTCAGGATGGAACAAGTGAGCCTGTGGCGTATGGCCCAGGCGTCGTCTCGCGGCCGCGGTAAGCCGACCCCGCCGCATCACCTCACAGTCAAAATCTTCATCGGGTTTGAATATGAATGTCCTAGAGGACACAGGCTGATGATGTCGTCCCCCACGAGTGCGGTGAGCGGGGGCTCGTGCGGGCGCGAGGCGGGGGCGCGGCTGGCGGCCAGCGACATGCCGCTGTACGTGCCCTGTCTGTGCTCCGTGCCCCTCCCCGCGCAGCTCACCCGGCTGCACGTCGTCACGCCCAAGGCTCCTGTACACGTCACGCTGGACCCCAGGGTGCAGCCGGTGCCCGGCGGGCCGATCTTCGTCCCGCAGCCCGTGGGCTCGCCGGCCATCAAGCTGTCGGCGTCCGCCTACTGGATCCTCCGCCTCCCGTTCGTGTACTGCGACGAGCGTGGCGCGCTGCCCCGCCCCCGCACCGCGCCCTCCTCCCCGCTGTCGCAAGGCGTTATACTAGCCCCACTGTTCGGGCTCGCCGACTGA
Protein
MKVFKLNDIPEFSKRERIVVVGIIGKSPHRYPNKTTPLLSAVQSQEIGIECHWDERRGVLFLHALTYLDTKHLSALAADLNENSKSTIKDADASNWLVASGELAIDSCKIIGLLFNLCHIVVLSSPTTVFDLGYLQLFKAIDAFRLEITNRTSVALSEAELGGTWISHGRCCCPRLLFHMRRAPVPLKRNPGALKRLEHSIEDQLYSILRKSRIITNVCAKALFAIPKNEEFVYISTDEEVCNARDVSVLVSGLVRMCGGAQPDRPAGERPGFAQFLQGHIDLAFAEGFDDNVGKYAMSTSFFELPSAESWRKAAEVLTPLYLEDPVQVDEGSEIGRTLFLILATDVRFSQARCAKILPIAQASYTEGMPAHYSSQHHAHKVKVALGVLETMARGPLAGGARARLQAACDVTWRSRSLCEALSLTGHPCIEPEHDGGTEHSSGVRYVSACACGRSKCSRDDPYTPRAANLAFYSLAADQCAHCRTLQAVAFPVFQPSTPTYKAASVKGAQSQEAYESSTVRETEGNSASGGEGGEGDESGGWSAPHALSPGSGGDDDEENKSPAAENRYVVPVTGDSNTDKMIRRQPSTTEYLPGMLHTASPAGLLPAYCSWSLVCVGASSLYSHSMGLPDHLQPGFLPHTNYLLPWDCAVRMEQVSLWRMAQASSRGRGKPTPPHHLTVKIFIGFEYECPRGHRLMMSSPTSAVSGGSCGREAGARLAASDMPLYVPCLCSVPLPAQLTRLHVVTPKAPVHVTLDPRVQPVPGGPIFVPQPVGSPAIKLSASAYWILRLPFVYCDERGALPRPRTAPSSPLSQGVILAPLFGLAD
Summary
Description
Involved in nonsense-mediated decay (NMD) of mRNAs containing premature stop codons. Probable component of kinase complex containing SMG1 and recruited to stalled ribosomes (By similarity).
Similarity
Belongs to the SMG8 family.
Keywords
Complete proteome
Nonsense-mediated mRNA decay
Reference proteome
Feature
chain Protein SMG8
Uniprot
A0A0N1IDX7
A0A194Q2A1
A0A2H1WU70
A0A212EMA1
E9I9E8
A0A151X4Z1
+ More
A0A0N0U4X6 K7IM59 E2C2F5 A0A232FCJ7 E2AYJ9 A0A1W4W9L7 A0A087ZNV5 A0A0C9QZU7 A0A195C6Y4 A0A2A3END4 A0A195FUN0 A0A026W6E2 A0A195DCR3 A0A2J7RA93 D6WLV3 A0A2S2PTR6 E0VLC2 A0A1Y1N4Z3 A0A0J7L2B1 A0A154NXU2 A0A310SNP8 N6UDI8 A0A195BBG7 U4UXZ9 A0A0P5CXL4 A0A2P8ZMG1 A0A0P5WWA2 A0A0P5X300 A0A158NAF5 A0A0N8ATD3 A0A0N8C2P8 A0A0P5H4Q0 A0A0P5YDR7 A0A0P5ZJL4 A0A0T6B2H2 B0W730 A0A182G2A5 Q17G65 E9G817 A0A0N8EK61 T1JDF4 A0A0P4YWM1 A0A0C9RFH3 A0A1I8PLZ8 A0A1A9UMJ9 A0A1I8N716 A0A0P5V3P1 A0A1B0FLQ3 A0A336LT62 A0A0L0C1U7 A0A0N8E839 C3XZ69 A0A0P6CIN9 R7VIE9 A0A1J1IMR3 A0A023F091 A0A0P4Z8J4 A0A0P5QLV7 A0A0N8CLP6 A0A093TCS1 H3ACG2 A0A1A8MTX0 A0A1A8NUJ9 A0A0Q3MF02 A0A1A7ZLS3 A0A1A8J720 A0A1A7Y2X7 A0A3B4FNS8 A0A1A7XGG0 A0A1A8JV23 A0A1A8D556 A0A1A8DD97 A0A1A8EVS1 A0A2I0LYQ4 A0A091M640 A0A3Q0H5P8 A0A091TP51 A0A3P8Q4I7 A0A3P9BTX5 A0A2I4CNZ2 A0A099ZE28 A0A093HMD8 R0K5X3 I3KL83 A0A287AXJ0 A0A091EKN1
A0A0N0U4X6 K7IM59 E2C2F5 A0A232FCJ7 E2AYJ9 A0A1W4W9L7 A0A087ZNV5 A0A0C9QZU7 A0A195C6Y4 A0A2A3END4 A0A195FUN0 A0A026W6E2 A0A195DCR3 A0A2J7RA93 D6WLV3 A0A2S2PTR6 E0VLC2 A0A1Y1N4Z3 A0A0J7L2B1 A0A154NXU2 A0A310SNP8 N6UDI8 A0A195BBG7 U4UXZ9 A0A0P5CXL4 A0A2P8ZMG1 A0A0P5WWA2 A0A0P5X300 A0A158NAF5 A0A0N8ATD3 A0A0N8C2P8 A0A0P5H4Q0 A0A0P5YDR7 A0A0P5ZJL4 A0A0T6B2H2 B0W730 A0A182G2A5 Q17G65 E9G817 A0A0N8EK61 T1JDF4 A0A0P4YWM1 A0A0C9RFH3 A0A1I8PLZ8 A0A1A9UMJ9 A0A1I8N716 A0A0P5V3P1 A0A1B0FLQ3 A0A336LT62 A0A0L0C1U7 A0A0N8E839 C3XZ69 A0A0P6CIN9 R7VIE9 A0A1J1IMR3 A0A023F091 A0A0P4Z8J4 A0A0P5QLV7 A0A0N8CLP6 A0A093TCS1 H3ACG2 A0A1A8MTX0 A0A1A8NUJ9 A0A0Q3MF02 A0A1A7ZLS3 A0A1A8J720 A0A1A7Y2X7 A0A3B4FNS8 A0A1A7XGG0 A0A1A8JV23 A0A1A8D556 A0A1A8DD97 A0A1A8EVS1 A0A2I0LYQ4 A0A091M640 A0A3Q0H5P8 A0A091TP51 A0A3P8Q4I7 A0A3P9BTX5 A0A2I4CNZ2 A0A099ZE28 A0A093HMD8 R0K5X3 I3KL83 A0A287AXJ0 A0A091EKN1
Pubmed
EMBL
KQ460650
KPJ13129.1
KQ459579
KPI99463.1
ODYU01011087
SOQ56603.1
+ More
AGBW02013858 OWR42615.1 GL761800 EFZ22785.1 KQ982544 KYQ55349.1 KQ435798 KOX73371.1 GL452124 EFN77905.1 NNAY01000419 OXU28536.1 GL443918 EFN61487.1 GBYB01000141 JAG69908.1 KQ978220 KYM96395.1 KZ288204 PBC33275.1 KQ981264 KYN43997.1 KK107374 QOIP01000010 EZA51583.1 RLU18184.1 KQ980989 KYN10700.1 NEVH01006576 PNF37753.1 KQ971343 EFA03396.2 GGMR01019617 MBY32236.1 DS235271 EEB14178.1 GEZM01013527 JAV92508.1 LBMM01001138 KMQ96836.1 KQ434781 KZC04496.1 KQ762329 OAD55967.1 APGK01032722 KB740848 ENN78691.1 KQ976528 KYM81878.1 KB632411 ERL95246.1 GDIP01164021 JAJ59381.1 PYGN01000016 PSN57691.1 GDIP01140632 GDIP01080681 GDIQ01030341 LRGB01000243 JAM23034.1 JAN64396.1 KZS20174.1 GDIP01078187 JAM25528.1 ADTU01001471 ADTU01001472 GDIQ01244795 JAK06930.1 GDIQ01120675 JAL31051.1 GDIQ01231442 JAK20283.1 GDIP01059240 JAM44475.1 GDIP01046280 JAM57435.1 LJIG01016131 KRT81522.1 DS231851 JXUM01138823 KQ568798 KXJ68880.1 CH477265 GL732534 EFX84763.1 GDIQ01018780 JAN75957.1 JH432105 GDIP01221972 JAJ01430.1 GBYB01011867 JAG81634.1 GDIP01105230 JAL98484.1 CCAG010020502 UFQT01000177 SSX21184.1 JRES01001119 KNC25374.1 GDIQ01052556 JAN42181.1 GG666475 EEN66633.1 GDIP01007916 JAM95799.1 AMQN01004377 KB293367 ELU16071.1 CVRI01000054 CRL00836.1 GBBI01003797 JAC14915.1 GDIP01216264 JAJ07138.1 GDIQ01112430 JAL39296.1 GDIP01119479 JAL84235.1 KL433323 KFW92204.1 AFYH01004161 HAEF01018867 HAEG01004270 SBR60026.1 HAEH01003992 HAEI01017105 SBR72745.1 LMAW01002498 KQK80936.1 HADY01004776 HAEJ01006173 SBP43261.1 HAED01019009 SBR05454.1 HADX01002103 SBP24335.1 HADW01015514 SBP16914.1 HAEE01003970 SBR23990.1 HADZ01022374 SBP86315.1 HAEA01002796 SBQ31276.1 HAEB01003417 HAEC01014014 SBQ49944.1 AKCR02000061 PKK22552.1 KK520115 KFP66094.1 KK461507 KFQ79401.1 KL892615 KGL79982.1 KL206227 KFV80172.1 KB742712 EOB05162.1 AERX01026576 KK718508 KFO57172.1
AGBW02013858 OWR42615.1 GL761800 EFZ22785.1 KQ982544 KYQ55349.1 KQ435798 KOX73371.1 GL452124 EFN77905.1 NNAY01000419 OXU28536.1 GL443918 EFN61487.1 GBYB01000141 JAG69908.1 KQ978220 KYM96395.1 KZ288204 PBC33275.1 KQ981264 KYN43997.1 KK107374 QOIP01000010 EZA51583.1 RLU18184.1 KQ980989 KYN10700.1 NEVH01006576 PNF37753.1 KQ971343 EFA03396.2 GGMR01019617 MBY32236.1 DS235271 EEB14178.1 GEZM01013527 JAV92508.1 LBMM01001138 KMQ96836.1 KQ434781 KZC04496.1 KQ762329 OAD55967.1 APGK01032722 KB740848 ENN78691.1 KQ976528 KYM81878.1 KB632411 ERL95246.1 GDIP01164021 JAJ59381.1 PYGN01000016 PSN57691.1 GDIP01140632 GDIP01080681 GDIQ01030341 LRGB01000243 JAM23034.1 JAN64396.1 KZS20174.1 GDIP01078187 JAM25528.1 ADTU01001471 ADTU01001472 GDIQ01244795 JAK06930.1 GDIQ01120675 JAL31051.1 GDIQ01231442 JAK20283.1 GDIP01059240 JAM44475.1 GDIP01046280 JAM57435.1 LJIG01016131 KRT81522.1 DS231851 JXUM01138823 KQ568798 KXJ68880.1 CH477265 GL732534 EFX84763.1 GDIQ01018780 JAN75957.1 JH432105 GDIP01221972 JAJ01430.1 GBYB01011867 JAG81634.1 GDIP01105230 JAL98484.1 CCAG010020502 UFQT01000177 SSX21184.1 JRES01001119 KNC25374.1 GDIQ01052556 JAN42181.1 GG666475 EEN66633.1 GDIP01007916 JAM95799.1 AMQN01004377 KB293367 ELU16071.1 CVRI01000054 CRL00836.1 GBBI01003797 JAC14915.1 GDIP01216264 JAJ07138.1 GDIQ01112430 JAL39296.1 GDIP01119479 JAL84235.1 KL433323 KFW92204.1 AFYH01004161 HAEF01018867 HAEG01004270 SBR60026.1 HAEH01003992 HAEI01017105 SBR72745.1 LMAW01002498 KQK80936.1 HADY01004776 HAEJ01006173 SBP43261.1 HAED01019009 SBR05454.1 HADX01002103 SBP24335.1 HADW01015514 SBP16914.1 HAEE01003970 SBR23990.1 HADZ01022374 SBP86315.1 HAEA01002796 SBQ31276.1 HAEB01003417 HAEC01014014 SBQ49944.1 AKCR02000061 PKK22552.1 KK520115 KFP66094.1 KK461507 KFQ79401.1 KL892615 KGL79982.1 KL206227 KFV80172.1 KB742712 EOB05162.1 AERX01026576 KK718508 KFO57172.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000075809
UP000053105
UP000002358
+ More
UP000008237 UP000215335 UP000000311 UP000192223 UP000005203 UP000078542 UP000242457 UP000078541 UP000053097 UP000279307 UP000078492 UP000235965 UP000007266 UP000009046 UP000036403 UP000076502 UP000019118 UP000078540 UP000030742 UP000245037 UP000076858 UP000005205 UP000002320 UP000069940 UP000249989 UP000008820 UP000000305 UP000095300 UP000078200 UP000095301 UP000092444 UP000037069 UP000001554 UP000014760 UP000183832 UP000008672 UP000051836 UP000261460 UP000053872 UP000189705 UP000265100 UP000265160 UP000192220 UP000053641 UP000053584 UP000005207 UP000008227 UP000052976
UP000008237 UP000215335 UP000000311 UP000192223 UP000005203 UP000078542 UP000242457 UP000078541 UP000053097 UP000279307 UP000078492 UP000235965 UP000007266 UP000009046 UP000036403 UP000076502 UP000019118 UP000078540 UP000030742 UP000245037 UP000076858 UP000005205 UP000002320 UP000069940 UP000249989 UP000008820 UP000000305 UP000095300 UP000078200 UP000095301 UP000092444 UP000037069 UP000001554 UP000014760 UP000183832 UP000008672 UP000051836 UP000261460 UP000053872 UP000189705 UP000265100 UP000265160 UP000192220 UP000053641 UP000053584 UP000005207 UP000008227 UP000052976
Interpro
IPR028802
SMG8
+ More
IPR019354 Smg8/Smg9
IPR036116 FN3_sf
IPR002423 Cpn60/TCP-1
IPR027484 PInositol-4-P-5-kinase_N
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR008401 Apc13p
IPR002498 PInositol-4-P-5-kinase_core
IPR011011 Znf_FYVE_PHD
IPR000306 Znf_FYVE
IPR000591 DEP_dom
IPR027409 GroEL-like_apical_dom_sf
IPR019354 Smg8/Smg9
IPR036116 FN3_sf
IPR002423 Cpn60/TCP-1
IPR027484 PInositol-4-P-5-kinase_N
IPR017455 Znf_FYVE-rel
IPR013083 Znf_RING/FYVE/PHD
IPR008401 Apc13p
IPR002498 PInositol-4-P-5-kinase_core
IPR011011 Znf_FYVE_PHD
IPR000306 Znf_FYVE
IPR000591 DEP_dom
IPR027409 GroEL-like_apical_dom_sf
Gene 3D
ProteinModelPortal
A0A0N1IDX7
A0A194Q2A1
A0A2H1WU70
A0A212EMA1
E9I9E8
A0A151X4Z1
+ More
A0A0N0U4X6 K7IM59 E2C2F5 A0A232FCJ7 E2AYJ9 A0A1W4W9L7 A0A087ZNV5 A0A0C9QZU7 A0A195C6Y4 A0A2A3END4 A0A195FUN0 A0A026W6E2 A0A195DCR3 A0A2J7RA93 D6WLV3 A0A2S2PTR6 E0VLC2 A0A1Y1N4Z3 A0A0J7L2B1 A0A154NXU2 A0A310SNP8 N6UDI8 A0A195BBG7 U4UXZ9 A0A0P5CXL4 A0A2P8ZMG1 A0A0P5WWA2 A0A0P5X300 A0A158NAF5 A0A0N8ATD3 A0A0N8C2P8 A0A0P5H4Q0 A0A0P5YDR7 A0A0P5ZJL4 A0A0T6B2H2 B0W730 A0A182G2A5 Q17G65 E9G817 A0A0N8EK61 T1JDF4 A0A0P4YWM1 A0A0C9RFH3 A0A1I8PLZ8 A0A1A9UMJ9 A0A1I8N716 A0A0P5V3P1 A0A1B0FLQ3 A0A336LT62 A0A0L0C1U7 A0A0N8E839 C3XZ69 A0A0P6CIN9 R7VIE9 A0A1J1IMR3 A0A023F091 A0A0P4Z8J4 A0A0P5QLV7 A0A0N8CLP6 A0A093TCS1 H3ACG2 A0A1A8MTX0 A0A1A8NUJ9 A0A0Q3MF02 A0A1A7ZLS3 A0A1A8J720 A0A1A7Y2X7 A0A3B4FNS8 A0A1A7XGG0 A0A1A8JV23 A0A1A8D556 A0A1A8DD97 A0A1A8EVS1 A0A2I0LYQ4 A0A091M640 A0A3Q0H5P8 A0A091TP51 A0A3P8Q4I7 A0A3P9BTX5 A0A2I4CNZ2 A0A099ZE28 A0A093HMD8 R0K5X3 I3KL83 A0A287AXJ0 A0A091EKN1
A0A0N0U4X6 K7IM59 E2C2F5 A0A232FCJ7 E2AYJ9 A0A1W4W9L7 A0A087ZNV5 A0A0C9QZU7 A0A195C6Y4 A0A2A3END4 A0A195FUN0 A0A026W6E2 A0A195DCR3 A0A2J7RA93 D6WLV3 A0A2S2PTR6 E0VLC2 A0A1Y1N4Z3 A0A0J7L2B1 A0A154NXU2 A0A310SNP8 N6UDI8 A0A195BBG7 U4UXZ9 A0A0P5CXL4 A0A2P8ZMG1 A0A0P5WWA2 A0A0P5X300 A0A158NAF5 A0A0N8ATD3 A0A0N8C2P8 A0A0P5H4Q0 A0A0P5YDR7 A0A0P5ZJL4 A0A0T6B2H2 B0W730 A0A182G2A5 Q17G65 E9G817 A0A0N8EK61 T1JDF4 A0A0P4YWM1 A0A0C9RFH3 A0A1I8PLZ8 A0A1A9UMJ9 A0A1I8N716 A0A0P5V3P1 A0A1B0FLQ3 A0A336LT62 A0A0L0C1U7 A0A0N8E839 C3XZ69 A0A0P6CIN9 R7VIE9 A0A1J1IMR3 A0A023F091 A0A0P4Z8J4 A0A0P5QLV7 A0A0N8CLP6 A0A093TCS1 H3ACG2 A0A1A8MTX0 A0A1A8NUJ9 A0A0Q3MF02 A0A1A7ZLS3 A0A1A8J720 A0A1A7Y2X7 A0A3B4FNS8 A0A1A7XGG0 A0A1A8JV23 A0A1A8D556 A0A1A8DD97 A0A1A8EVS1 A0A2I0LYQ4 A0A091M640 A0A3Q0H5P8 A0A091TP51 A0A3P8Q4I7 A0A3P9BTX5 A0A2I4CNZ2 A0A099ZE28 A0A093HMD8 R0K5X3 I3KL83 A0A287AXJ0 A0A091EKN1
PDB
5NKM
E-value=2.79309e-07,
Score=134
Ontologies
GO
PANTHER
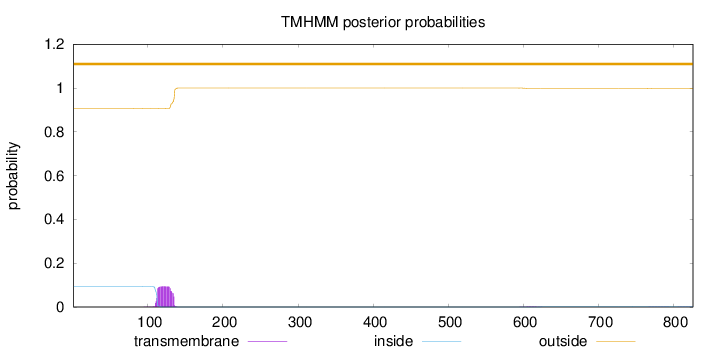
Topology
Length:
826
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.1366
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.09402
outside
1 - 826
Population Genetic Test Statistics
Pi
191.870549
Theta
186.84699
Tajima's D
0.222287
CLR
0.183748
CSRT
0.432328383580821
Interpretation
Uncertain
