Pre Gene Modal
BGIBMGA013598
Annotation
PREDICTED:_AP-1_complex_subunit_sigma-2_isoform_X2_[Plutella_xylostella]
Full name
AP complex subunit sigma
Location in the cell
Cytoplasmic Reliability : 1.573 Golgi Reliability : 1.131
Sequence
CDS
ATGATGCAGTTTATGCTATTATTTAGCCGTCAAGGCAAATTGAGGCTTCAGAAGTGGTACGTGGCTCATCCAGATAAGCTAAAGAAGAAAATAACTAGAGAATTGATTACCACAGTGCTAGCTCGAAAACCCAAAATGTGTTCTTTCCTTGAATGGAAAGATGTTAAAGTTGTGTATAAAAGATATGCATCACTATATTTCTGCTGTGCTATGGAACAAGAAGACAATGAGCTGCTGACCTTGGAGTTGATCCATCGCTATGTAGAACTGCTGGACAAGTATTTTGGCAGTGTTTGTGAACTGGACATCATCTTTAACTTTGAGAAAGCATACTTCATACTTGATGAACTGGTTCTTGGTGGAGAGCTACAAGAAACCAGTAAGAAGAATGTCCTGAAAGCCATCGCTGCCCAGGATCTTCTGCAGGAGGAGGAGACACCGCAGGGCTTCTTCGAAGACCACGGCCTCGGATAA
Protein
MMQFMLLFSRQGKLRLQKWYVAHPDKLKKKITRELITTVLARKPKMCSFLEWKDVKVVYKRYASLYFCCAMEQEDNELLTLELIHRYVELLDKYFGSVCELDIIFNFEKAYFILDELVLGGELQETSKKNVLKAIAAQDLLQEEETPQGFFEDHGLG
Summary
Similarity
Belongs to the adaptor complexes small subunit family.
Feature
chain AP complex subunit sigma
Uniprot
A0A2W1BGV6
A0A1E1WES3
Q2F5U0
A0A212EM28
A0A1B6ECU6
A0A1Y1L6K7
+ More
A0A0T6BGV2 A0A1S3DCN4 A0A1Y1L365 V9ID69 E2ACP9 A0A3L8DBF2 A0A1B6GGE6 A0A293MVQ7 A0A087TIH9 A0A1E1XJY2 A0A1E1X0U2 D6WD27 J3JU90 L7M339 A0A154NWD7 K7J0N4 A0A0M8ZXP9 A0A158P3M2 A0A1B6KLT0 A0A146L2I4 A0A1Q3G3A0 Q17MI7 D1FQ10 U5EWK2 A0A0K8RIZ9 A0A2L2Y7R0 A0A1D2MBX0 J9K426 A0A1L8DVX1 A0A2H8TYJ3 A0A2S2QZR5 A0A0L8FH31 A0A1B0CQ08 K1PU11 A0A1S3I1J8 A0A2A3EPS6 A0A2M4CU87 A0A2M4C217 A0A182ND36 F5HLJ6 A0A182FR30 D3TQF6 A0A0K8UMB1 A0A034W3P0 V3ZV51 A0A0L0C7K9 T1PAD7 C4N170 A0A1L8EB84 A0A1I8PS45 A0A1W4UKB4 A0A3B0JQT1 Q298M4 B4G4B6 B4PL44 B3M313 B4N9E5 Q9VCF4 A0A1E1W4B9 B4M0G7 B4K6U6 A0A0M4ESQ2 A0A131XW95 I6WHE4 B4QSD8 B4HG13 B3P8B4 A0A0P6I3U8 A0A0P6B4D9 B4JT85 A0A0N1PJ42 Q9GQM8 A0A158QBG9 A0A182E4D6 A0A1B6CCP1 A0A183XFD6 A0A2H1W2Z7 A0A0M3IN99 F1LHA0 S4P3U1 A0A1S3DBC7 A0A0R3RVP7 A0A1S0U2C4 A0A0J9XRQ0 A0A0R3Q4S4 A0A1E1WAE4 A0A222AJE1 A0A139WMJ9 A0A1Y1L5E3 G0MXN1 A0A2G5TJZ2 O16369 A0A0B2VNX0 I6VE66
A0A0T6BGV2 A0A1S3DCN4 A0A1Y1L365 V9ID69 E2ACP9 A0A3L8DBF2 A0A1B6GGE6 A0A293MVQ7 A0A087TIH9 A0A1E1XJY2 A0A1E1X0U2 D6WD27 J3JU90 L7M339 A0A154NWD7 K7J0N4 A0A0M8ZXP9 A0A158P3M2 A0A1B6KLT0 A0A146L2I4 A0A1Q3G3A0 Q17MI7 D1FQ10 U5EWK2 A0A0K8RIZ9 A0A2L2Y7R0 A0A1D2MBX0 J9K426 A0A1L8DVX1 A0A2H8TYJ3 A0A2S2QZR5 A0A0L8FH31 A0A1B0CQ08 K1PU11 A0A1S3I1J8 A0A2A3EPS6 A0A2M4CU87 A0A2M4C217 A0A182ND36 F5HLJ6 A0A182FR30 D3TQF6 A0A0K8UMB1 A0A034W3P0 V3ZV51 A0A0L0C7K9 T1PAD7 C4N170 A0A1L8EB84 A0A1I8PS45 A0A1W4UKB4 A0A3B0JQT1 Q298M4 B4G4B6 B4PL44 B3M313 B4N9E5 Q9VCF4 A0A1E1W4B9 B4M0G7 B4K6U6 A0A0M4ESQ2 A0A131XW95 I6WHE4 B4QSD8 B4HG13 B3P8B4 A0A0P6I3U8 A0A0P6B4D9 B4JT85 A0A0N1PJ42 Q9GQM8 A0A158QBG9 A0A182E4D6 A0A1B6CCP1 A0A183XFD6 A0A2H1W2Z7 A0A0M3IN99 F1LHA0 S4P3U1 A0A1S3DBC7 A0A0R3RVP7 A0A1S0U2C4 A0A0J9XRQ0 A0A0R3Q4S4 A0A1E1WAE4 A0A222AJE1 A0A139WMJ9 A0A1Y1L5E3 G0MXN1 A0A2G5TJZ2 O16369 A0A0B2VNX0 I6VE66
Pubmed
28756777
22118469
28004739
20798317
30249741
29209593
+ More
28503490 18362917 19820115 22516182 23537049 25576852 20075255 21347285 26823975 17510324 26561354 27289101 22992520 12364791 14747013 17210077 20353571 25348373 23254933 26108605 25315136 19576987 15632085 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26354079 10954086 22919073 26850696 21685128 23622113 17885136 9851916
28503490 18362917 19820115 22516182 23537049 25576852 20075255 21347285 26823975 17510324 26561354 27289101 22992520 12364791 14747013 17210077 20353571 25348373 23254933 26108605 25315136 19576987 15632085 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26354079 10954086 22919073 26850696 21685128 23622113 17885136 9851916
EMBL
KZ150071
PZC74048.1
GDQN01005637
JAT85417.1
DQ311333
JQ824197
+ More
ABD36277.1 AFN25811.1 AGBW02013960 OWR42511.1 GEDC01031524 GEDC01001555 JAS05774.1 JAS35743.1 GEZM01066281 JAV68010.1 LJIG01000418 KRT86522.1 GEZM01066279 GEZM01066277 JAV68014.1 JR038824 AEY58426.1 GL438568 EFN68793.1 QOIP01000010 RLU17835.1 GECZ01008406 JAS61363.1 GFWV01019313 MAA44041.1 KK115358 KFM64918.1 GFAA01003807 JAT99627.1 GFAC01006298 JAT92890.1 KQ971312 EEZ98327.1 APGK01020369 BT126803 KB740193 KB631602 AEE61765.1 ENN81149.1 ERL84545.1 GACK01006298 JAA58736.1 KQ434773 KZC03986.1 KQ435801 KOX73250.1 ADTU01008261 ADTU01008262 GEBQ01027565 JAT12412.1 GDHC01017222 JAQ01407.1 GFDL01000765 JAV34280.1 CH477205 EAT47937.1 EZ419910 ACZ28265.1 GANO01001379 JAB58492.1 GADI01003309 JAA70499.1 IAAA01009710 LAA04188.1 LJIJ01002000 ODM90384.1 ABLF02020332 GFDF01003515 JAV10569.1 GFXV01007165 MBW18970.1 GGMS01013961 MBY83164.1 KQ431871 KOF63003.1 AJWK01022902 AJWK01022903 AJWK01022904 AJWK01022905 JH818022 EKC27782.1 KZ288197 PBC33717.1 GGFL01004230 MBW68408.1 GGFJ01010194 MBW59335.1 AAAB01008898 EGK97129.1 EZ423658 ADD19934.1 GDHF01024809 JAI27505.1 GAKP01010554 JAC48398.1 KB203083 ESO86445.1 JRES01000809 KNC28241.1 KA645697 AFP60326.1 EZ048843 ACN69135.1 GFDG01002986 JAV15813.1 OUUW01000007 SPP82752.1 CM000070 EAL27931.2 CH479179 EDW24464.1 CM000160 EDW99029.2 CH902617 EDV42413.2 CH964232 EDW80578.2 AE014297 AY061172 KX531465 AAF56212.2 AAL28720.1 ANY27275.1 GDQN01009323 JAT81731.1 CH940650 EDW68346.2 CH933806 EDW14212.1 CP012526 ALC47810.1 GEFM01004497 JAP71299.1 JQ824199 AFN25813.1 CM000364 EDX14137.1 CH480815 EDW43406.1 CH954182 EDV53938.1 GDIQ01026458 JAN68279.1 GDIP01020235 JAM83480.1 CH916373 EDV94975.1 KQ460650 KPJ13133.1 AF228054 AAG43051.1 UXUI01009904 VDD94484.1 UYRW01000489 VDK67262.1 GEDC01026107 JAS11191.1 UYWW01000080 VDM07176.1 ODYU01005868 SOQ47192.1 JI219945 ADY49504.1 GAIX01006294 JAA86266.1 JH712310 EFO23363.1 LN856891 CDP93665.1 UZAG01000402 VDO08269.1 GDQN01007139 JAT83915.1 KY563925 KY609774 ASO76371.1 ASO76489.1 KYB29045.1 GEZM01066280 JAV68011.1 GL379819 EGT47188.1 PDUG01000005 PIC27553.1 BX284605 CCD64132.1 JPKZ01001231 UYWY01024532 KHN83074.1 VDM48853.1 JQ824198 AFN25812.1
ABD36277.1 AFN25811.1 AGBW02013960 OWR42511.1 GEDC01031524 GEDC01001555 JAS05774.1 JAS35743.1 GEZM01066281 JAV68010.1 LJIG01000418 KRT86522.1 GEZM01066279 GEZM01066277 JAV68014.1 JR038824 AEY58426.1 GL438568 EFN68793.1 QOIP01000010 RLU17835.1 GECZ01008406 JAS61363.1 GFWV01019313 MAA44041.1 KK115358 KFM64918.1 GFAA01003807 JAT99627.1 GFAC01006298 JAT92890.1 KQ971312 EEZ98327.1 APGK01020369 BT126803 KB740193 KB631602 AEE61765.1 ENN81149.1 ERL84545.1 GACK01006298 JAA58736.1 KQ434773 KZC03986.1 KQ435801 KOX73250.1 ADTU01008261 ADTU01008262 GEBQ01027565 JAT12412.1 GDHC01017222 JAQ01407.1 GFDL01000765 JAV34280.1 CH477205 EAT47937.1 EZ419910 ACZ28265.1 GANO01001379 JAB58492.1 GADI01003309 JAA70499.1 IAAA01009710 LAA04188.1 LJIJ01002000 ODM90384.1 ABLF02020332 GFDF01003515 JAV10569.1 GFXV01007165 MBW18970.1 GGMS01013961 MBY83164.1 KQ431871 KOF63003.1 AJWK01022902 AJWK01022903 AJWK01022904 AJWK01022905 JH818022 EKC27782.1 KZ288197 PBC33717.1 GGFL01004230 MBW68408.1 GGFJ01010194 MBW59335.1 AAAB01008898 EGK97129.1 EZ423658 ADD19934.1 GDHF01024809 JAI27505.1 GAKP01010554 JAC48398.1 KB203083 ESO86445.1 JRES01000809 KNC28241.1 KA645697 AFP60326.1 EZ048843 ACN69135.1 GFDG01002986 JAV15813.1 OUUW01000007 SPP82752.1 CM000070 EAL27931.2 CH479179 EDW24464.1 CM000160 EDW99029.2 CH902617 EDV42413.2 CH964232 EDW80578.2 AE014297 AY061172 KX531465 AAF56212.2 AAL28720.1 ANY27275.1 GDQN01009323 JAT81731.1 CH940650 EDW68346.2 CH933806 EDW14212.1 CP012526 ALC47810.1 GEFM01004497 JAP71299.1 JQ824199 AFN25813.1 CM000364 EDX14137.1 CH480815 EDW43406.1 CH954182 EDV53938.1 GDIQ01026458 JAN68279.1 GDIP01020235 JAM83480.1 CH916373 EDV94975.1 KQ460650 KPJ13133.1 AF228054 AAG43051.1 UXUI01009904 VDD94484.1 UYRW01000489 VDK67262.1 GEDC01026107 JAS11191.1 UYWW01000080 VDM07176.1 ODYU01005868 SOQ47192.1 JI219945 ADY49504.1 GAIX01006294 JAA86266.1 JH712310 EFO23363.1 LN856891 CDP93665.1 UZAG01000402 VDO08269.1 GDQN01007139 JAT83915.1 KY563925 KY609774 ASO76371.1 ASO76489.1 KYB29045.1 GEZM01066280 JAV68011.1 GL379819 EGT47188.1 PDUG01000005 PIC27553.1 BX284605 CCD64132.1 JPKZ01001231 UYWY01024532 KHN83074.1 VDM48853.1 JQ824198 AFN25812.1
Proteomes
UP000007151
UP000079169
UP000000311
UP000279307
UP000054359
UP000007266
+ More
UP000019118 UP000030742 UP000076502 UP000002358 UP000053105 UP000005205 UP000008820 UP000094527 UP000007819 UP000053454 UP000092461 UP000005408 UP000085678 UP000242457 UP000075884 UP000007062 UP000069272 UP000030746 UP000037069 UP000095301 UP000095300 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000007801 UP000007798 UP000000803 UP000008792 UP000009192 UP000092553 UP000000304 UP000001292 UP000008711 UP000001070 UP000053240 UP000038041 UP000274131 UP000077448 UP000271087 UP000093561 UP000270924 UP000036681 UP000050640 UP000006672 UP000050602 UP000008068 UP000230233 UP000001940 UP000031036 UP000050794 UP000267007
UP000019118 UP000030742 UP000076502 UP000002358 UP000053105 UP000005205 UP000008820 UP000094527 UP000007819 UP000053454 UP000092461 UP000005408 UP000085678 UP000242457 UP000075884 UP000007062 UP000069272 UP000030746 UP000037069 UP000095301 UP000095300 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000007801 UP000007798 UP000000803 UP000008792 UP000009192 UP000092553 UP000000304 UP000001292 UP000008711 UP000001070 UP000053240 UP000038041 UP000274131 UP000077448 UP000271087 UP000093561 UP000270924 UP000036681 UP000050640 UP000006672 UP000050602 UP000008068 UP000230233 UP000001940 UP000031036 UP000050794 UP000267007
Interpro
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
A0A2W1BGV6
A0A1E1WES3
Q2F5U0
A0A212EM28
A0A1B6ECU6
A0A1Y1L6K7
+ More
A0A0T6BGV2 A0A1S3DCN4 A0A1Y1L365 V9ID69 E2ACP9 A0A3L8DBF2 A0A1B6GGE6 A0A293MVQ7 A0A087TIH9 A0A1E1XJY2 A0A1E1X0U2 D6WD27 J3JU90 L7M339 A0A154NWD7 K7J0N4 A0A0M8ZXP9 A0A158P3M2 A0A1B6KLT0 A0A146L2I4 A0A1Q3G3A0 Q17MI7 D1FQ10 U5EWK2 A0A0K8RIZ9 A0A2L2Y7R0 A0A1D2MBX0 J9K426 A0A1L8DVX1 A0A2H8TYJ3 A0A2S2QZR5 A0A0L8FH31 A0A1B0CQ08 K1PU11 A0A1S3I1J8 A0A2A3EPS6 A0A2M4CU87 A0A2M4C217 A0A182ND36 F5HLJ6 A0A182FR30 D3TQF6 A0A0K8UMB1 A0A034W3P0 V3ZV51 A0A0L0C7K9 T1PAD7 C4N170 A0A1L8EB84 A0A1I8PS45 A0A1W4UKB4 A0A3B0JQT1 Q298M4 B4G4B6 B4PL44 B3M313 B4N9E5 Q9VCF4 A0A1E1W4B9 B4M0G7 B4K6U6 A0A0M4ESQ2 A0A131XW95 I6WHE4 B4QSD8 B4HG13 B3P8B4 A0A0P6I3U8 A0A0P6B4D9 B4JT85 A0A0N1PJ42 Q9GQM8 A0A158QBG9 A0A182E4D6 A0A1B6CCP1 A0A183XFD6 A0A2H1W2Z7 A0A0M3IN99 F1LHA0 S4P3U1 A0A1S3DBC7 A0A0R3RVP7 A0A1S0U2C4 A0A0J9XRQ0 A0A0R3Q4S4 A0A1E1WAE4 A0A222AJE1 A0A139WMJ9 A0A1Y1L5E3 G0MXN1 A0A2G5TJZ2 O16369 A0A0B2VNX0 I6VE66
A0A0T6BGV2 A0A1S3DCN4 A0A1Y1L365 V9ID69 E2ACP9 A0A3L8DBF2 A0A1B6GGE6 A0A293MVQ7 A0A087TIH9 A0A1E1XJY2 A0A1E1X0U2 D6WD27 J3JU90 L7M339 A0A154NWD7 K7J0N4 A0A0M8ZXP9 A0A158P3M2 A0A1B6KLT0 A0A146L2I4 A0A1Q3G3A0 Q17MI7 D1FQ10 U5EWK2 A0A0K8RIZ9 A0A2L2Y7R0 A0A1D2MBX0 J9K426 A0A1L8DVX1 A0A2H8TYJ3 A0A2S2QZR5 A0A0L8FH31 A0A1B0CQ08 K1PU11 A0A1S3I1J8 A0A2A3EPS6 A0A2M4CU87 A0A2M4C217 A0A182ND36 F5HLJ6 A0A182FR30 D3TQF6 A0A0K8UMB1 A0A034W3P0 V3ZV51 A0A0L0C7K9 T1PAD7 C4N170 A0A1L8EB84 A0A1I8PS45 A0A1W4UKB4 A0A3B0JQT1 Q298M4 B4G4B6 B4PL44 B3M313 B4N9E5 Q9VCF4 A0A1E1W4B9 B4M0G7 B4K6U6 A0A0M4ESQ2 A0A131XW95 I6WHE4 B4QSD8 B4HG13 B3P8B4 A0A0P6I3U8 A0A0P6B4D9 B4JT85 A0A0N1PJ42 Q9GQM8 A0A158QBG9 A0A182E4D6 A0A1B6CCP1 A0A183XFD6 A0A2H1W2Z7 A0A0M3IN99 F1LHA0 S4P3U1 A0A1S3DBC7 A0A0R3RVP7 A0A1S0U2C4 A0A0J9XRQ0 A0A0R3Q4S4 A0A1E1WAE4 A0A222AJE1 A0A139WMJ9 A0A1Y1L5E3 G0MXN1 A0A2G5TJZ2 O16369 A0A0B2VNX0 I6VE66
PDB
1W63
E-value=1.22454e-61,
Score=594
Ontologies
GO
PANTHER
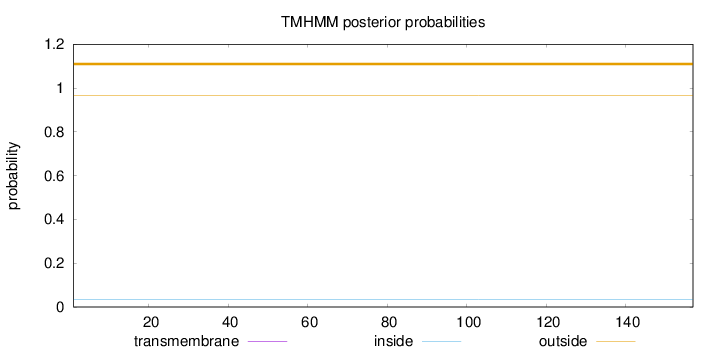
Topology
Length:
157
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00045
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03429
outside
1 - 157
Population Genetic Test Statistics
Pi
240.55019
Theta
206.108096
Tajima's D
1.015168
CLR
0.002878
CSRT
0.658817059147043
Interpretation
Uncertain
