Gene
KWMTBOMO03247
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.944
Sequence
CDS
ATGACAGTTAAAAAAAGGAAAAGGGTTTTGTCGGCGATGCCCCATTCCACATTTAATGTGGATTTCAGCAATGTAAGGGGCCTACATAGTAACCTCGACGCCGTACACCACCATCTTGAGACGGCGCAGCCTGCCCTCCTTTTTCTGACGGAGACACAGATATCTGCTCCGGATGACACTTCGTACCTTGAATACCCCGGCTACGTATTGGAGCACAACTTCCTGCGTAAAGCCGGGGTGTGTGTATTCGTCCGGGCTGATGTCTGTTGTCGCCGTCTACGAGGCCTCGAACAACGGGACCTGTCCATCTTGTGGCTGCGCGTAGATCACGGGGGTTGTACCCGAGTCTACGCGTGCCTGTACAGGTCCCACAGCAGTGATGCAGGTTCAGCTCTGATAGAGCATGTGCAAGAGGGGACTAACCGCGTGCTTGAGCAGTACCCATCTGCGGAGGTGGTGGTTCTTGGAGACTTTAACGCTCACCACCAAGAGTGGTTGGGGTCCAGAACCACTGACCTCCCGGGTCGGGCTGCCTACGATTTCGCCCTGGCCTACGGCTTCTCCCAGCTGGTGACAGAGCCCACCCGTGTCCCAGATATTGAGGGGCACGAGCCCTCTCTGTTGGACCTTCTGCTGACCACCGATCCAGCCGGATACAGTGTGGTGGTCGACGCTCCACTTGGATCGTCTGATCACTGCCTTATCCGTGCTGCGGTACCGATCTCTCGTCCTGGTCGTCGAATGATGACCGGGTATCGAAGAGTTTGGCGGTATATGTCAGCAGATTGGGATGGGTTGCGTGAATTTTACGCATCCTACCCATGGGGGCGGCTCTGCTTTTCCTCTGGTGATCCTGACGTCTGTGCGGACCGACTTAAAAACGTGGTGCTTCGGGGGATGGAACTGTTCATTCCTTCTTCCGAGGTGCCCGTCGGGGGTCGCAGTGGACCCTGGTATAACAATGCCAGTCGGGACGCTGCACGCCTCAAGCGGTCCGCATACGCGGCATGGAATGATGCCAGGAGACGCCAGGATCCTAATACCTCAGAGGAAAGGCGGAAATTTAACGCCGCTTCTAGGTCCTATAAGAGGGCTATTGCCAGGGCGAAGTCGGAGCACGTTGCCAGAATTGGCGAGCGACTGAAGAGCTATCCCGCCGGGAGCCGTGCTTTCTGGTCGCTCGCTAAGGCTGCAGAAGGTAACTTCTGCAGGTCTAGTCTCCCACCACTACGCAAGTCTGATGACAGTCTGGCCCATAGTGCGAAAGAGAAGGCTGACCTTCTGGTCAAACTCTTCGCCTCGAACTCGACTGTGGACGACGGGGGTGCCACACCACCGAACATCCCCCGGTGTGACAGTTCCCTGCCGGAGATCTGCTTTACACAGTGTGCAGTCAGGCGGGAACTCCGACTCCTGGACGTCCATAAGTCGAGTGGGCCGGACGGCATCCCTGCTGTGGTTCTGAAAAGGTGA
Protein
MTVKKRKRVLSAMPHSTFNVDFSNVRGLHSNLDAVHHHLETAQPALLFLTETQISAPDDTSYLEYPGYVLEHNFLRKAGVCVFVRADVCCRRLRGLEQRDLSILWLRVDHGGCTRVYACLYRSHSSDAGSALIEHVQEGTNRVLEQYPSAEVVVLGDFNAHHQEWLGSRTTDLPGRAAYDFALAYGFSQLVTEPTRVPDIEGHEPSLLDLLLTTDPAGYSVVVDAPLGSSDHCLIRAAVPISRPGRRMMTGYRRVWRYMSADWDGLREFYASYPWGRLCFSSGDPDVCADRLKNVVLRGMELFIPSSEVPVGGRSGPWYNNASRDAARLKRSAYAAWNDARRRQDPNTSEERRKFNAASRSYKRAIARAKSEHVARIGERLKSYPAGSRAFWSLAKAAEGNFCRSSLPPLRKSDDSLAHSAKEKADLLVKLFASNSTVDDGGATPPNIPRCDSSLPEICFTQCAVRRELRLLDVHKSSGPDGIPAVVLKR
Summary
Uniprot
Q6UV17
A0A3D5S1K1
T2M4I6
A0A2H9T4N9
A0A0P4VPK5
A0A0P4VVV8
+ More
W4YYU9 A0A2H9T4U7 A0A147BMN4 A0A147BMW1 A0A2H9T3K2 D6WYM1 W4XUR2 D7GYL9 A0A147BIC7 A0A0P4VX79 W4YEI4 Q8WS60 A0A354GIR8 A0A2H9T4F1 W4YYW9 A0A2H2IW73 W4YXS6 A0A2H9T441 A0A147BBB4 A0A2B4RAD6 A0A2B4RBK9 D6WYM3 A0A2H9T4Y4 W4YK32 A0A2H9T462 A0A2H9T5P0 A0A2B4RB46 D6WYM2 A0A2B4RCD2 A0A2B4S7G9 A0A2B4R707 A0A210QEX7 A0A147BJ61 A0A147BQS9 A0A2B4RML3 A0A2B4R7V5 A0A131XUP0 A0A2B4RWS1 W4Z6B2 A0A0X3PTK8 A0A147BJG7 A0A131Y4U2 A0A147BLG6 A0A147BJJ5 E3MBS8 W4Y4X9 A0A147BNC9 A0A2B4SQF8 A0A1E1XH11 A0A2B4R4J4 A0A2B4SCV9 A0A2B4RDR7 A0A147BQ97 W4Z6B0 D7ELW9 W4Z6B1 E3LJB3 A0A1E1XG83 D7GYM0 D7EM24 L7MD77 A0A147AJ39 A0A2B4T296 A0A3M0L7V8 A0A147BM11 L7LSU8 R4GDE5 A0A147BLZ0 A0A147BI04 A0A1X7VCX9 A0A147BJE7 L7MD44
W4YYU9 A0A2H9T4U7 A0A147BMN4 A0A147BMW1 A0A2H9T3K2 D6WYM1 W4XUR2 D7GYL9 A0A147BIC7 A0A0P4VX79 W4YEI4 Q8WS60 A0A354GIR8 A0A2H9T4F1 W4YYW9 A0A2H2IW73 W4YXS6 A0A2H9T441 A0A147BBB4 A0A2B4RAD6 A0A2B4RBK9 D6WYM3 A0A2H9T4Y4 W4YK32 A0A2H9T462 A0A2H9T5P0 A0A2B4RB46 D6WYM2 A0A2B4RCD2 A0A2B4S7G9 A0A2B4R707 A0A210QEX7 A0A147BJ61 A0A147BQS9 A0A2B4RML3 A0A2B4R7V5 A0A131XUP0 A0A2B4RWS1 W4Z6B2 A0A0X3PTK8 A0A147BJG7 A0A131Y4U2 A0A147BLG6 A0A147BJJ5 E3MBS8 W4Y4X9 A0A147BNC9 A0A2B4SQF8 A0A1E1XH11 A0A2B4R4J4 A0A2B4SCV9 A0A2B4RDR7 A0A147BQ97 W4Z6B0 D7ELW9 W4Z6B1 E3LJB3 A0A1E1XG83 D7GYM0 D7EM24 L7MD77 A0A147AJ39 A0A2B4T296 A0A3M0L7V8 A0A147BM11 L7LSU8 R4GDE5 A0A147BLZ0 A0A147BI04 A0A1X7VCX9 A0A147BJE7 L7MD44
Pubmed
EMBL
AY359886
AAQ57129.1
DPOC01000269
HCX22453.1
HAAD01000593
CDG66825.1
+ More
NSIT01000234 PJE78200.1 GDRN01111278 JAI56803.1 GDRN01111277 JAI56804.1 AAGJ04114595 NSIT01000225 PJE78229.1 GEGO01003370 JAR92034.1 GEGO01003274 JAR92130.1 NSIT01000383 PJE77769.1 KQ971357 EFA08424.1 AAGJ04060226 GG695857 EFA13460.1 GEGO01004865 JAR90539.1 GDRN01111326 JAI56798.1 AAGJ04103823 AH011247 AAL40415.1 DNUT01000434 HBI40906.1 NSIT01000271 PJE78081.1 AAGJ04015404 AAGJ04111627 NSIT01000300 PJE77993.1 GEGO01007351 JAR88053.1 LSMT01000899 PFX13773.1 LSMT01000767 PFX14526.1 EFA08429.1 NSIT01000218 PJE78267.1 AAGJ04029969 NSIT01000294 PJE78013.1 NSIT01000164 PJE78524.1 LSMT01000907 PFX13730.1 EFA08423.1 LSMT01000661 PFX15311.1 LSMT01000174 PFX24547.1 LSMT01001623 PFX12055.1 NEDP02004020 OWF47171.1 GEGO01004596 JAR90808.1 GEGO01002277 JAR93127.1 LSMT01000412 PFX18406.1 PFX13731.1 GEFM01005848 JAP69948.1 LSMT01000295 PFX21010.1 AAGJ04167750 GEEE01008367 JAP54858.1 GEGO01004477 JAR90927.1 GEFM01002301 JAP73495.1 GEGO01003776 JAR91628.1 GEGO01004441 JAR90963.1 DS268433 EFO97776.1 AAGJ04110620 GEGO01003111 JAR92293.1 LSMT01000046 PFX30727.1 GFAC01000857 JAT98331.1 LSMT01001965 PFX11729.1 LSMT01000121 PFX26660.1 LSMT01000769 PFX14518.1 GEGO01002473 JAR92931.1 DS497938 EFA12429.1 DS268409 EFO94840.1 GFAC01000913 JAT98275.1 EFA13459.1 DS497963 EFA12534.1 GACK01003890 JAA61144.1 GCES01007896 JAR78427.1 LSMT01000001 PFX34902.1 QRBI01000093 RMC21672.1 GEGO01003567 JAR91837.1 GACK01009888 JAA55146.1 GEGO01003594 JAR91810.1 GEGO01005479 JAR89925.1 GEGO01004518 JAR90886.1 GACK01003881 JAA61153.1
NSIT01000234 PJE78200.1 GDRN01111278 JAI56803.1 GDRN01111277 JAI56804.1 AAGJ04114595 NSIT01000225 PJE78229.1 GEGO01003370 JAR92034.1 GEGO01003274 JAR92130.1 NSIT01000383 PJE77769.1 KQ971357 EFA08424.1 AAGJ04060226 GG695857 EFA13460.1 GEGO01004865 JAR90539.1 GDRN01111326 JAI56798.1 AAGJ04103823 AH011247 AAL40415.1 DNUT01000434 HBI40906.1 NSIT01000271 PJE78081.1 AAGJ04015404 AAGJ04111627 NSIT01000300 PJE77993.1 GEGO01007351 JAR88053.1 LSMT01000899 PFX13773.1 LSMT01000767 PFX14526.1 EFA08429.1 NSIT01000218 PJE78267.1 AAGJ04029969 NSIT01000294 PJE78013.1 NSIT01000164 PJE78524.1 LSMT01000907 PFX13730.1 EFA08423.1 LSMT01000661 PFX15311.1 LSMT01000174 PFX24547.1 LSMT01001623 PFX12055.1 NEDP02004020 OWF47171.1 GEGO01004596 JAR90808.1 GEGO01002277 JAR93127.1 LSMT01000412 PFX18406.1 PFX13731.1 GEFM01005848 JAP69948.1 LSMT01000295 PFX21010.1 AAGJ04167750 GEEE01008367 JAP54858.1 GEGO01004477 JAR90927.1 GEFM01002301 JAP73495.1 GEGO01003776 JAR91628.1 GEGO01004441 JAR90963.1 DS268433 EFO97776.1 AAGJ04110620 GEGO01003111 JAR92293.1 LSMT01000046 PFX30727.1 GFAC01000857 JAT98331.1 LSMT01001965 PFX11729.1 LSMT01000121 PFX26660.1 LSMT01000769 PFX14518.1 GEGO01002473 JAR92931.1 DS497938 EFA12429.1 DS268409 EFO94840.1 GFAC01000913 JAT98275.1 EFA13459.1 DS497963 EFA12534.1 GACK01003890 JAA61144.1 GCES01007896 JAR78427.1 LSMT01000001 PFX34902.1 QRBI01000093 RMC21672.1 GEGO01003567 JAR91837.1 GACK01009888 JAA55146.1 GEGO01003594 JAR91810.1 GEGO01005479 JAR89925.1 GEGO01004518 JAR90886.1 GACK01003881 JAA61153.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR017850 Alkaline_phosphatase_core_sf
IPR000917 Sulfatase_N
IPR024607 Sulfatase_CS
IPR021896 Transposase_37
IPR035983 Hect_E3_ubiquitin_ligase
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR022812 Dynamin_SF
IPR008266 Tyr_kinase_AS
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR027417 P-loop_NTPase
IPR011604 Exonuc_phg/RecB_C
IPR008042 Retrotrans_Pao
IPR036034 PDZ_sf
IPR001478 PDZ
IPR001965 Znf_PHD
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR017850 Alkaline_phosphatase_core_sf
IPR000917 Sulfatase_N
IPR024607 Sulfatase_CS
IPR021896 Transposase_37
IPR035983 Hect_E3_ubiquitin_ligase
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR022812 Dynamin_SF
IPR008266 Tyr_kinase_AS
IPR000719 Prot_kinase_dom
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR027417 P-loop_NTPase
IPR011604 Exonuc_phg/RecB_C
IPR008042 Retrotrans_Pao
IPR036034 PDZ_sf
IPR001478 PDZ
IPR001965 Znf_PHD
SUPFAM
ProteinModelPortal
Q6UV17
A0A3D5S1K1
T2M4I6
A0A2H9T4N9
A0A0P4VPK5
A0A0P4VVV8
+ More
W4YYU9 A0A2H9T4U7 A0A147BMN4 A0A147BMW1 A0A2H9T3K2 D6WYM1 W4XUR2 D7GYL9 A0A147BIC7 A0A0P4VX79 W4YEI4 Q8WS60 A0A354GIR8 A0A2H9T4F1 W4YYW9 A0A2H2IW73 W4YXS6 A0A2H9T441 A0A147BBB4 A0A2B4RAD6 A0A2B4RBK9 D6WYM3 A0A2H9T4Y4 W4YK32 A0A2H9T462 A0A2H9T5P0 A0A2B4RB46 D6WYM2 A0A2B4RCD2 A0A2B4S7G9 A0A2B4R707 A0A210QEX7 A0A147BJ61 A0A147BQS9 A0A2B4RML3 A0A2B4R7V5 A0A131XUP0 A0A2B4RWS1 W4Z6B2 A0A0X3PTK8 A0A147BJG7 A0A131Y4U2 A0A147BLG6 A0A147BJJ5 E3MBS8 W4Y4X9 A0A147BNC9 A0A2B4SQF8 A0A1E1XH11 A0A2B4R4J4 A0A2B4SCV9 A0A2B4RDR7 A0A147BQ97 W4Z6B0 D7ELW9 W4Z6B1 E3LJB3 A0A1E1XG83 D7GYM0 D7EM24 L7MD77 A0A147AJ39 A0A2B4T296 A0A3M0L7V8 A0A147BM11 L7LSU8 R4GDE5 A0A147BLZ0 A0A147BI04 A0A1X7VCX9 A0A147BJE7 L7MD44
W4YYU9 A0A2H9T4U7 A0A147BMN4 A0A147BMW1 A0A2H9T3K2 D6WYM1 W4XUR2 D7GYL9 A0A147BIC7 A0A0P4VX79 W4YEI4 Q8WS60 A0A354GIR8 A0A2H9T4F1 W4YYW9 A0A2H2IW73 W4YXS6 A0A2H9T441 A0A147BBB4 A0A2B4RAD6 A0A2B4RBK9 D6WYM3 A0A2H9T4Y4 W4YK32 A0A2H9T462 A0A2H9T5P0 A0A2B4RB46 D6WYM2 A0A2B4RCD2 A0A2B4S7G9 A0A2B4R707 A0A210QEX7 A0A147BJ61 A0A147BQS9 A0A2B4RML3 A0A2B4R7V5 A0A131XUP0 A0A2B4RWS1 W4Z6B2 A0A0X3PTK8 A0A147BJG7 A0A131Y4U2 A0A147BLG6 A0A147BJJ5 E3MBS8 W4Y4X9 A0A147BNC9 A0A2B4SQF8 A0A1E1XH11 A0A2B4R4J4 A0A2B4SCV9 A0A2B4RDR7 A0A147BQ97 W4Z6B0 D7ELW9 W4Z6B1 E3LJB3 A0A1E1XG83 D7GYM0 D7EM24 L7MD77 A0A147AJ39 A0A2B4T296 A0A3M0L7V8 A0A147BM11 L7LSU8 R4GDE5 A0A147BLZ0 A0A147BI04 A0A1X7VCX9 A0A147BJE7 L7MD44
Ontologies
GO
PANTHER
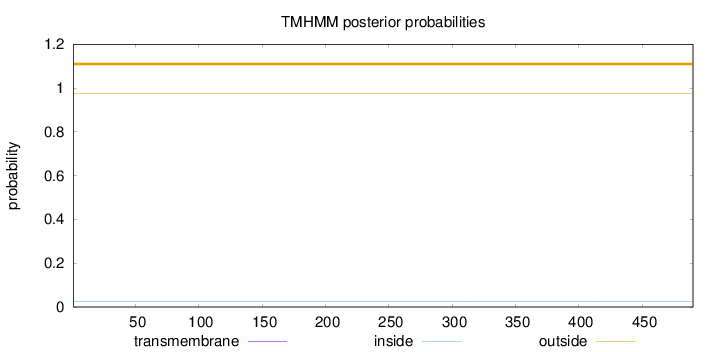
Topology
Length:
490
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0024
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.02623
outside
1 - 490
Population Genetic Test Statistics
Pi
12.673347
Theta
101.678657
Tajima's D
0
CLR
0.046161
CSRT
0.36023198840058
Interpretation
Uncertain
