Pre Gene Modal
BGIBMGA013614
Annotation
PREDICTED:_xylulose_kinase_isoform_X2_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 1.338
Sequence
CDS
ATGTTTGAGGAATCAAAGAATAAAAGTTTTTTAGGGTTTGATTTTAGTACACAGAGGTTAAAAGCACTAGTAATCAATGAAGACTACAATGTACTCCACGAGGCTGATGTTGAATTTGATGTTGACTTGCCTGAGTTTAGGACCGCAGGGGGTGTGGTGCGCGGCGAGAAGGGTGAGGTAACAGCACCTCCCCTTTTATGGGTGAAAGCATTAGACATGGTTTTGGACCGACTAATTGTAGCTGGACTAGACTTCTCCACTGTGTATGCTATATCTGGAGCAGCTCAGCAACACGGTTCTGTGTGGTGGTCGAAAGATGCCGAAGCGAAATTAGGCAGTTTGTCATCAGACGACTTCTTGCACACTCAGTTGGCGTCCGCATTTTTGTGCGACTCCCCAGTTTGGATGGACTCTAGTACGTCTGCCGACTGCAGGGCTCTGGAGGAAGCCATGGGTGGACCAGAGGAATTAGCTAAAATCACAGGATCTAGGGCGTACGAACGTTTTACCGGGCCACAGATTCGTAAAATGTTCCGTACTCGGCCCCGGGTGTACCAGGCCGCGCCCCGGATATCCCTGGTGTCCTCGTTCGCGTGCTCGCTGTTCCTGGGCAAGATCGCGCCCATCGACCTGGCGGACGGATCCGGAATGAACCTTCTGGACATTAGGACTATGCAATGGGACGAACGGGCACTGAAGGCATGCGGGGACGAGTCGCTGATCCACAAACTGGGTGCGGCAGTCCCCAGCGCGAGCGACGTCGGGCCGGTGTCCGCCTACTTCGTGCAGCGGTACGGCTTCAGGCCCGACTGCCGCGTGGTGGCCTTCACCGGGGACAACTGCTCTGCCCTGGCCGGCCTTAGACTTCGTTCGGGTTGGATAGGGCTCAGCCTGGGCACCAGCGACACCCTGCTGCTCGGCCTCACGGAGCCCGCGGCCCCGGCGGCCGGCCACGTGCTCGTGGGGCCGACTCCCGACGCTCCGTACATGGCGTTACTGTGCTTCGCCAACGGCTCGCTGACGAGGCAGAACCACAGGGACCGGCTGGTGGGCCCGAGCTGGGAGGCGTTTAATGAGCTCCTCAGGTCGACCGTCCGAGGGAATATGGGATTCATGGGAGTGTACTACGACACAGCAGAGATAGTGCCGCGCCGCGCCGCCGCCCGCCTGCTGCAGGCCAGCGGAGGCCGCGCCGTGCAGGCCGGGGCTCCGGCGGCGGAGGCCCGCGCGCTGCTGGAGGGGCAGGCGCTCGCCCGTAGAGCGCACGCCGAGGACCTGCACTTCACTATCGGCCCCTCGTCCAAGCTGATAGCGACCGGGGGCGCCGCCGCCAACAAAGAGTTCCTCCAGATATTTGCTGACGTCTTCAATGTTCCCGTGTACGTACTGGGCGAGCACGGCAACGCGGCTCTGCTGGGGGCCGCCGTCCGCGCCGCCGCGGTGTGGTCCCGGGACGCCGCCGTGTCGCTGCCAGGCTCGGAGCCCACCCTGACCCCGGTGGCGACGCCCTACCCGGACCACGACGCCATCTACACGCCGATGCTGGCGCGCTACCGGCAGATCATCCAGGAGATGCCCCGGCTGCACTGA
Protein
MFEESKNKSFLGFDFSTQRLKALVINEDYNVLHEADVEFDVDLPEFRTAGGVVRGEKGEVTAPPLLWVKALDMVLDRLIVAGLDFSTVYAISGAAQQHGSVWWSKDAEAKLGSLSSDDFLHTQLASAFLCDSPVWMDSSTSADCRALEEAMGGPEELAKITGSRAYERFTGPQIRKMFRTRPRVYQAAPRISLVSSFACSLFLGKIAPIDLADGSGMNLLDIRTMQWDERALKACGDESLIHKLGAAVPSASDVGPVSAYFVQRYGFRPDCRVVAFTGDNCSALAGLRLRSGWIGLSLGTSDTLLLGLTEPAAPAAGHVLVGPTPDAPYMALLCFANGSLTRQNHRDRLVGPSWEAFNELLRSTVRGNMGFMGVYYDTAEIVPRRAAARLLQASGGRAVQAGAPAAEARALLEGQALARRAHAEDLHFTIGPSSKLIATGGAAANKEFLQIFADVFNVPVYVLGEHGNAALLGAAVRAAAVWSRDAAVSLPGSEPTLTPVATPYPDHDAIYTPMLARYRQIIQEMPRLH
Summary
Uniprot
H9JVQ1
A0A2H1VYC4
A0A2A4J9J3
A0A2W1BQP7
A0A194Q2B0
A0A212EM00
+ More
A0A0L7LMU4 K7J6G5 A0A1B6E7I8 A0A088AG69 A0A1Y1LDQ7 A0A1B6GE18 A0A2A3EPV2 A0A1B6HDA3 D6WB21 E9IJM8 A0A232F7Y7 A0A154NWP8 E2BMY1 A0A2J7QUC0 F4WW65 N6UHC6 U4U067 A0A2J7QUC3 A0A195F1C4 A0A0C9QWV2 A0A067QYR5 A0A026WJE6 A0A195B2I5 J9JX02 A0A0V0G6J5 A0A0L7QLZ4 A0A0A9ZD51 A0A023F3A2 A0A146LER5 A0A2R7X6D8 A0A0K8SH32 A0A0P4VPP5 T1I2X7 A0A310SE62 A0A1S3IQ97 A0A0C9REN4 A0A195CZQ4 V5HSE7 A0A0P4WBD4 B7PI19 A0A1S3J5G9 A0A2B4S7Z1 T1JTD0 H2YXB8 F6SAH8 A0A1W5B6C6 A0A0M4EFW6 A0A3P8YRX4 E2ATB3 C3Z072 A0A2G8KWS9 A0A293MWT2 A0A1D5PKV6 A0A0M8ZXQ8 U3JWE7 A0A3M6TSC9 G3WAV7 A0A151PCY7 A0A1Q3FP83 Q16TL1 A0A2J7QUC4 B0X8P6 Q16I25 A0A1D2N0C0 A0A1V4KLI3 A0A2P2IEH1 A0A218UYI7 A0A1E1X4A6 A0A182RCJ0 A0A182J585 A0A182WTP3 A0A182URQ6 B3M2B9 A0A182IAE1 Q7QEY9 A0A2I4D1A8 U6CRC7 A0A210QRP6 Q7T342 G1KM28 M3W9P2 A0A2Y9KVQ3 A0A2U3WSQ2 A0A1B6JM97 A0A287DG07 A0A182JNX0 A7RMC9 A0A146XXZ2 A0A1U7QMY5 F1PI40 F1RRB3 A0A0P5NVY6 A0A182PJD7 A0A3B3RDG4
A0A0L7LMU4 K7J6G5 A0A1B6E7I8 A0A088AG69 A0A1Y1LDQ7 A0A1B6GE18 A0A2A3EPV2 A0A1B6HDA3 D6WB21 E9IJM8 A0A232F7Y7 A0A154NWP8 E2BMY1 A0A2J7QUC0 F4WW65 N6UHC6 U4U067 A0A2J7QUC3 A0A195F1C4 A0A0C9QWV2 A0A067QYR5 A0A026WJE6 A0A195B2I5 J9JX02 A0A0V0G6J5 A0A0L7QLZ4 A0A0A9ZD51 A0A023F3A2 A0A146LER5 A0A2R7X6D8 A0A0K8SH32 A0A0P4VPP5 T1I2X7 A0A310SE62 A0A1S3IQ97 A0A0C9REN4 A0A195CZQ4 V5HSE7 A0A0P4WBD4 B7PI19 A0A1S3J5G9 A0A2B4S7Z1 T1JTD0 H2YXB8 F6SAH8 A0A1W5B6C6 A0A0M4EFW6 A0A3P8YRX4 E2ATB3 C3Z072 A0A2G8KWS9 A0A293MWT2 A0A1D5PKV6 A0A0M8ZXQ8 U3JWE7 A0A3M6TSC9 G3WAV7 A0A151PCY7 A0A1Q3FP83 Q16TL1 A0A2J7QUC4 B0X8P6 Q16I25 A0A1D2N0C0 A0A1V4KLI3 A0A2P2IEH1 A0A218UYI7 A0A1E1X4A6 A0A182RCJ0 A0A182J585 A0A182WTP3 A0A182URQ6 B3M2B9 A0A182IAE1 Q7QEY9 A0A2I4D1A8 U6CRC7 A0A210QRP6 Q7T342 G1KM28 M3W9P2 A0A2Y9KVQ3 A0A2U3WSQ2 A0A1B6JM97 A0A287DG07 A0A182JNX0 A7RMC9 A0A146XXZ2 A0A1U7QMY5 F1PI40 F1RRB3 A0A0P5NVY6 A0A182PJD7 A0A3B3RDG4
Pubmed
19121390
28756777
26354079
22118469
26227816
20075255
+ More
28004739 18362917 19820115 21282665 28648823 20798317 21719571 23537049 24845553 24508170 30249741 25401762 25474469 26823975 27129103 25765539 12481130 15114417 25069045 18563158 29023486 15592404 30382153 21709235 22293439 17510324 27289101 28503490 17994087 12364791 14747013 17210077 28812685 17975172 17615350 16341006 29240929
28004739 18362917 19820115 21282665 28648823 20798317 21719571 23537049 24845553 24508170 30249741 25401762 25474469 26823975 27129103 25765539 12481130 15114417 25069045 18563158 29023486 15592404 30382153 21709235 22293439 17510324 27289101 28503490 17994087 12364791 14747013 17210077 28812685 17975172 17615350 16341006 29240929
EMBL
BABH01032328
BABH01032329
ODYU01005188
SOQ45803.1
NWSH01002255
PCG68797.1
+ More
KZ150071 PZC74043.1 KQ459579 KPI99473.1 AGBW02013960 OWR42515.1 JTDY01000534 KOB76762.1 GEDC01003396 JAS33902.1 GEZM01061442 JAV70480.1 GECZ01024305 GECZ01009102 JAS45464.1 JAS60667.1 KZ288197 PBC33720.1 GECU01035043 JAS72663.1 KQ971311 EEZ98930.1 GL763877 EFZ19200.1 NNAY01000777 OXU26563.1 KQ434773 KZC04011.1 GL449382 EFN82885.1 NEVH01011186 PNF32173.1 GL888404 EGI61536.1 APGK01035437 KB740925 ENN78037.1 KB631924 ERL87269.1 PNF32175.1 KQ981864 KYN34375.1 GBYB01005152 JAG74919.1 KK853184 KDR10119.1 KK107185 QOIP01000012 EZA55801.1 RLU15750.1 KQ976662 KYM78414.1 ABLF02014747 ABLF02014748 ABLF02014751 ABLF02014752 ABLF02052021 GECL01002752 JAP03372.1 KQ414914 KOC59536.1 GBHO01001230 JAG42374.1 GBBI01003036 JAC15676.1 GDHC01011978 JAQ06651.1 KK857495 PTY27231.1 GBRD01013387 GBRD01013386 JAG52439.1 GDKW01002244 JAI54351.1 ACPB03020332 KQ771002 OAD52555.1 GBYB01011607 JAG81374.1 KQ977115 KYN05619.1 GANP01005566 JAB78902.1 GDRN01071491 JAI63674.1 ABJB010109450 DS716413 EEC06241.1 LSMT01000136 PFX26011.1 CAEY01000474 EAAA01002410 CP012526 ALC47141.1 GL442545 EFN63358.1 GG666567 EEN54131.1 MRZV01000328 PIK52463.1 GFWV01018897 MAA43625.1 AADN05000033 KQ435801 KOX73255.1 AGTO01020343 RCHS01003049 RMX44188.1 AEFK01085896 AEFK01085897 AEFK01085898 AEFK01085899 AEFK01085900 AEFK01085901 AEFK01085902 AEFK01085903 AEFK01085904 AEFK01085905 AKHW03000487 KYO46966.1 GFDL01005723 JAV29322.1 CH477646 EAT37839.1 PNF32179.1 DS232495 EDS42638.1 CH478115 EAT33917.1 LJIJ01000330 ODM98719.1 LSYS01002888 OPJ85271.1 IACF01006820 LAB72403.1 MUZQ01000098 OWK58480.1 GFAC01005096 JAT94092.1 CH902617 EDV42310.1 APCN01001201 AAAB01008846 EAA06371.4 HAAF01001394 CCP73220.1 NEDP02002268 OWF51404.1 BC053262 AAH53262.1 AANG04001816 GECU01007310 JAT00397.1 AGTP01006788 DS469520 EDO47320.1 GCES01040261 JAR46062.1 AAEX03013491 AAEX03013492 AEMK02000085 GDIQ01136925 JAL14801.1
KZ150071 PZC74043.1 KQ459579 KPI99473.1 AGBW02013960 OWR42515.1 JTDY01000534 KOB76762.1 GEDC01003396 JAS33902.1 GEZM01061442 JAV70480.1 GECZ01024305 GECZ01009102 JAS45464.1 JAS60667.1 KZ288197 PBC33720.1 GECU01035043 JAS72663.1 KQ971311 EEZ98930.1 GL763877 EFZ19200.1 NNAY01000777 OXU26563.1 KQ434773 KZC04011.1 GL449382 EFN82885.1 NEVH01011186 PNF32173.1 GL888404 EGI61536.1 APGK01035437 KB740925 ENN78037.1 KB631924 ERL87269.1 PNF32175.1 KQ981864 KYN34375.1 GBYB01005152 JAG74919.1 KK853184 KDR10119.1 KK107185 QOIP01000012 EZA55801.1 RLU15750.1 KQ976662 KYM78414.1 ABLF02014747 ABLF02014748 ABLF02014751 ABLF02014752 ABLF02052021 GECL01002752 JAP03372.1 KQ414914 KOC59536.1 GBHO01001230 JAG42374.1 GBBI01003036 JAC15676.1 GDHC01011978 JAQ06651.1 KK857495 PTY27231.1 GBRD01013387 GBRD01013386 JAG52439.1 GDKW01002244 JAI54351.1 ACPB03020332 KQ771002 OAD52555.1 GBYB01011607 JAG81374.1 KQ977115 KYN05619.1 GANP01005566 JAB78902.1 GDRN01071491 JAI63674.1 ABJB010109450 DS716413 EEC06241.1 LSMT01000136 PFX26011.1 CAEY01000474 EAAA01002410 CP012526 ALC47141.1 GL442545 EFN63358.1 GG666567 EEN54131.1 MRZV01000328 PIK52463.1 GFWV01018897 MAA43625.1 AADN05000033 KQ435801 KOX73255.1 AGTO01020343 RCHS01003049 RMX44188.1 AEFK01085896 AEFK01085897 AEFK01085898 AEFK01085899 AEFK01085900 AEFK01085901 AEFK01085902 AEFK01085903 AEFK01085904 AEFK01085905 AKHW03000487 KYO46966.1 GFDL01005723 JAV29322.1 CH477646 EAT37839.1 PNF32179.1 DS232495 EDS42638.1 CH478115 EAT33917.1 LJIJ01000330 ODM98719.1 LSYS01002888 OPJ85271.1 IACF01006820 LAB72403.1 MUZQ01000098 OWK58480.1 GFAC01005096 JAT94092.1 CH902617 EDV42310.1 APCN01001201 AAAB01008846 EAA06371.4 HAAF01001394 CCP73220.1 NEDP02002268 OWF51404.1 BC053262 AAH53262.1 AANG04001816 GECU01007310 JAT00397.1 AGTP01006788 DS469520 EDO47320.1 GCES01040261 JAR46062.1 AAEX03013491 AAEX03013492 AEMK02000085 GDIQ01136925 JAL14801.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000002358
+ More
UP000005203 UP000242457 UP000007266 UP000215335 UP000076502 UP000008237 UP000235965 UP000007755 UP000019118 UP000030742 UP000078541 UP000027135 UP000053097 UP000279307 UP000078540 UP000007819 UP000053825 UP000015103 UP000085678 UP000078542 UP000001555 UP000225706 UP000015104 UP000007875 UP000008144 UP000092553 UP000265140 UP000000311 UP000001554 UP000230750 UP000000539 UP000053105 UP000016665 UP000275408 UP000007648 UP000050525 UP000008820 UP000002320 UP000094527 UP000190648 UP000197619 UP000075900 UP000075880 UP000076407 UP000075903 UP000007801 UP000075840 UP000007062 UP000192220 UP000242188 UP000001646 UP000011712 UP000248482 UP000245340 UP000005215 UP000075881 UP000001593 UP000265000 UP000189706 UP000002254 UP000008227 UP000075885 UP000261540
UP000005203 UP000242457 UP000007266 UP000215335 UP000076502 UP000008237 UP000235965 UP000007755 UP000019118 UP000030742 UP000078541 UP000027135 UP000053097 UP000279307 UP000078540 UP000007819 UP000053825 UP000015103 UP000085678 UP000078542 UP000001555 UP000225706 UP000015104 UP000007875 UP000008144 UP000092553 UP000265140 UP000000311 UP000001554 UP000230750 UP000000539 UP000053105 UP000016665 UP000275408 UP000007648 UP000050525 UP000008820 UP000002320 UP000094527 UP000190648 UP000197619 UP000075900 UP000075880 UP000076407 UP000075903 UP000007801 UP000075840 UP000007062 UP000192220 UP000242188 UP000001646 UP000011712 UP000248482 UP000245340 UP000005215 UP000075881 UP000001593 UP000265000 UP000189706 UP000002254 UP000008227 UP000075885 UP000261540
PRIDE
Interpro
ProteinModelPortal
H9JVQ1
A0A2H1VYC4
A0A2A4J9J3
A0A2W1BQP7
A0A194Q2B0
A0A212EM00
+ More
A0A0L7LMU4 K7J6G5 A0A1B6E7I8 A0A088AG69 A0A1Y1LDQ7 A0A1B6GE18 A0A2A3EPV2 A0A1B6HDA3 D6WB21 E9IJM8 A0A232F7Y7 A0A154NWP8 E2BMY1 A0A2J7QUC0 F4WW65 N6UHC6 U4U067 A0A2J7QUC3 A0A195F1C4 A0A0C9QWV2 A0A067QYR5 A0A026WJE6 A0A195B2I5 J9JX02 A0A0V0G6J5 A0A0L7QLZ4 A0A0A9ZD51 A0A023F3A2 A0A146LER5 A0A2R7X6D8 A0A0K8SH32 A0A0P4VPP5 T1I2X7 A0A310SE62 A0A1S3IQ97 A0A0C9REN4 A0A195CZQ4 V5HSE7 A0A0P4WBD4 B7PI19 A0A1S3J5G9 A0A2B4S7Z1 T1JTD0 H2YXB8 F6SAH8 A0A1W5B6C6 A0A0M4EFW6 A0A3P8YRX4 E2ATB3 C3Z072 A0A2G8KWS9 A0A293MWT2 A0A1D5PKV6 A0A0M8ZXQ8 U3JWE7 A0A3M6TSC9 G3WAV7 A0A151PCY7 A0A1Q3FP83 Q16TL1 A0A2J7QUC4 B0X8P6 Q16I25 A0A1D2N0C0 A0A1V4KLI3 A0A2P2IEH1 A0A218UYI7 A0A1E1X4A6 A0A182RCJ0 A0A182J585 A0A182WTP3 A0A182URQ6 B3M2B9 A0A182IAE1 Q7QEY9 A0A2I4D1A8 U6CRC7 A0A210QRP6 Q7T342 G1KM28 M3W9P2 A0A2Y9KVQ3 A0A2U3WSQ2 A0A1B6JM97 A0A287DG07 A0A182JNX0 A7RMC9 A0A146XXZ2 A0A1U7QMY5 F1PI40 F1RRB3 A0A0P5NVY6 A0A182PJD7 A0A3B3RDG4
A0A0L7LMU4 K7J6G5 A0A1B6E7I8 A0A088AG69 A0A1Y1LDQ7 A0A1B6GE18 A0A2A3EPV2 A0A1B6HDA3 D6WB21 E9IJM8 A0A232F7Y7 A0A154NWP8 E2BMY1 A0A2J7QUC0 F4WW65 N6UHC6 U4U067 A0A2J7QUC3 A0A195F1C4 A0A0C9QWV2 A0A067QYR5 A0A026WJE6 A0A195B2I5 J9JX02 A0A0V0G6J5 A0A0L7QLZ4 A0A0A9ZD51 A0A023F3A2 A0A146LER5 A0A2R7X6D8 A0A0K8SH32 A0A0P4VPP5 T1I2X7 A0A310SE62 A0A1S3IQ97 A0A0C9REN4 A0A195CZQ4 V5HSE7 A0A0P4WBD4 B7PI19 A0A1S3J5G9 A0A2B4S7Z1 T1JTD0 H2YXB8 F6SAH8 A0A1W5B6C6 A0A0M4EFW6 A0A3P8YRX4 E2ATB3 C3Z072 A0A2G8KWS9 A0A293MWT2 A0A1D5PKV6 A0A0M8ZXQ8 U3JWE7 A0A3M6TSC9 G3WAV7 A0A151PCY7 A0A1Q3FP83 Q16TL1 A0A2J7QUC4 B0X8P6 Q16I25 A0A1D2N0C0 A0A1V4KLI3 A0A2P2IEH1 A0A218UYI7 A0A1E1X4A6 A0A182RCJ0 A0A182J585 A0A182WTP3 A0A182URQ6 B3M2B9 A0A182IAE1 Q7QEY9 A0A2I4D1A8 U6CRC7 A0A210QRP6 Q7T342 G1KM28 M3W9P2 A0A2Y9KVQ3 A0A2U3WSQ2 A0A1B6JM97 A0A287DG07 A0A182JNX0 A7RMC9 A0A146XXZ2 A0A1U7QMY5 F1PI40 F1RRB3 A0A0P5NVY6 A0A182PJD7 A0A3B3RDG4
PDB
4BC5
E-value=1.2012e-115,
Score=1067
Ontologies
PATHWAY
GO
Topology
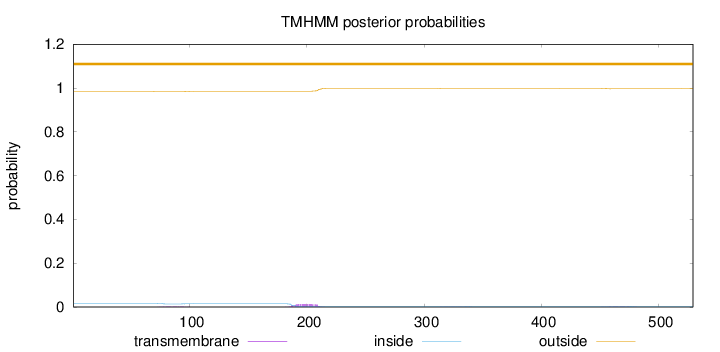
Length:
529
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.379500000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01545
outside
1 - 529
Population Genetic Test Statistics
Pi
244.975208
Theta
171.507131
Tajima's D
2.29801
CLR
0.373641
CSRT
0.923153842307885
Interpretation
Uncertain
