Gene
KWMTBOMO03245 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013594
Annotation
Splicing_factor_3A_subunit_1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.248 Nuclear Reliability : 1.798
Sequence
CDS
ATGCCATCTATAGATGTAATGCTACCACCGGGGCAGTCGTTAGAGATCCCGGAAGATCCGCCTCCACCACCCCCTCCAGTCATTGGCATTATTTATCCACCACCTGAAGTCAGAAATATTGTTGATAAAACTGCCAGCTTTGTGGCTCGTAATGGTCCTGAATTTGAAGCTAGGATCAGACAGAATGAGCTGGGCAATCCAAAGTTCAACTTCCTTAATTCAGGGGATCCCTACCATGCTTACTATCAGCATAAAGTCAGAGAGATCAAAGAGGGAAAAGTTCCCGAACCAACTGCACCACCAACTATGCCATCAGGACAGCAACGAATGGGTCTCGCTCCAGCCACAGCGGCCCGCCAGCAGGAGCTCCTCAAGGCAGCTGCTCCTGCAGAACTGCCTCCGCCGCGCGACCCACCACCAGATTTCGAATTTATCGCTGATCCACCGTCTATATCGGCTCTGGAGCTCGACATAGTGAAATTAACCGCGCAATTTGTCGCCAGAAACGGTCGTCAGTTCTTAACGGATTTAATGAAGAAAGAGGAGCGTAATCATCAGTTTGATTTTCTTCGTCCCCAGCATTCACTGTTCCAATACTTCACGCGTCTGCTTGAGCAGTACACGAAGGTGTTGCTGCCGCCAAAAGAACTAGTATCAAAGCTGAATGCTGAGATACGCAGCGGGATATTAGAACAAGCTCGCAGTCGCGCTGCCTGGCACGCGCACCAAGCCAAGCGCAAGGCCGCTGACGAGGCCAAAATTGAGAAAGAGAGGCTCGCGTATGCCTCCATAGACTGGCACGATTTTGTCGTTGTTGAAACTGTAGATTACCCCGCAGGGGAACCCGGAGACTTCCCTCCTCCGACCACTCCACTAGAAGTCGGAGCCCGCGTCCTCGCCCAAGAGAGGGGTGAAGATATAATACGGAACCAGAATGACGACGATGATACTGAGATGCAAATTGAATCCGAATCGGAATCGGAGTCTGATGAGGAACTTGCGGAAATGGAGGATCGAACTCAGTCCCATCGGCCGGACGATAACCGCGTGCAAGACATGGAAGAGGAAAGCTCGACTTCGGAGTCCGAGACGGAGCAAGACGCCCCGCGCGCGGACGAGGCGCCCCTCCCGCCGCGCCCCGACCGCGTCGTCGTCAAGAAGTACAACCCCAAGGAGGCGAAGCCCCGCGCCGCCGCCGGGGACGAGTGGCTCGTGTCGCCCATCACCGGCGAGAAGATCCCCGCCAGCAAGGTGGCCGAGCACGTGCGCATCGGGCTGCTGGACCCGCGCTGGCTGGAGCAGCGGGACCGCGCCGCCGCCGAGCGCACCGACCGGGACGAGGCGCTCGCCCCCGGGGCCGCCATCGAGGCTTCGCTTAAACAGTTGGCGGAGCGTCGTACTGACATCTTCGGTGTAGGGGACGAGGAAACGACCATCGGTAAGAAGATCGGCGAGGAGGAGAAGAAGCGCGACGAGCGCGTGACGTGGGACGGGCACACCTCCAGCGTGGAGGCCGCCACCCGCGCCGCCAGGGCGCACATCACGCTCGAGGACCAGATCCAGCAGATACACAAGGTTAAAGGTCTACTCCCCGACGAGGAGAAAGAGAAAATAGGTCCGAAGCCAGTGACCGGGGCAGCCCAGAGCCGGGCGGGGCGTCCCCCCGCGCCCCCCTCTGCCCCCCGCGCTGCTCCGCCTCCCCCGCCCCCGGCACCGCCTGTGCTGCTGCAGCCGATGTCCTCGCTAATGGTCCTGCCCCCCATGGCGCCCCGACCTCCGCTCATACCGGCTCCAGTGGGTGCAGTACCACCGCCGGCGTTCAACTTCGGAGCGGTAGTCCCTCCGGGCACAGTGGCGGAGCCCGAGGACGAGGGCCCGGCCGCTAAGAGAGCCCGCACCGAGGACGCGCTGGAGCCCGAGGGCGCCTGGCTCGCGCGCCACCCCGGCGCAGTCCCCCTGCAGGTGTCGTTGCCGGCGGCGCCTGAGCGCGCGGAGTGGCGGCTGGACGGGCGCGCGGTGCCGCTGACGCTGCCGCTGTCGGCCGCCGTCGCCGAGCTCAAGGCCATGCTGCAGCGCGCCACCAACATGCCCACCGCCAAGCAGAAGCTGCACTACGAGGGTCTGTTCTTCAAGGATACTAACAGCTTGGCGTACTACAACGTGCCCCCGGGTGCCGTCATACAGCTACAGATCAAGGAACGAGGCGGTAGAAAGAAATAA
Protein
MPSIDVMLPPGQSLEIPEDPPPPPPPVIGIIYPPPEVRNIVDKTASFVARNGPEFEARIRQNELGNPKFNFLNSGDPYHAYYQHKVREIKEGKVPEPTAPPTMPSGQQRMGLAPATAARQQELLKAAAPAELPPPRDPPPDFEFIADPPSISALELDIVKLTAQFVARNGRQFLTDLMKKEERNHQFDFLRPQHSLFQYFTRLLEQYTKVLLPPKELVSKLNAEIRSGILEQARSRAAWHAHQAKRKAADEAKIEKERLAYASIDWHDFVVVETVDYPAGEPGDFPPPTTPLEVGARVLAQERGEDIIRNQNDDDDTEMQIESESESESDEELAEMEDRTQSHRPDDNRVQDMEEESSTSESETEQDAPRADEAPLPPRPDRVVVKKYNPKEAKPRAAAGDEWLVSPITGEKIPASKVAEHVRIGLLDPRWLEQRDRAAAERTDRDEALAPGAAIEASLKQLAERRTDIFGVGDEETTIGKKIGEEEKKRDERVTWDGHTSSVEAATRAARAHITLEDQIQQIHKVKGLLPDEEKEKIGPKPVTGAAQSRAGRPPAPPSAPRAAPPPPPPAPPVLLQPMSSLMVLPPMAPRPPLIPAPVGAVPPPAFNFGAVVPPGTVAEPEDEGPAAKRARTEDALEPEGAWLARHPGAVPLQVSLPAAPERAEWRLDGRAVPLTLPLSAAVAELKAMLQRATNMPTAKQKLHYEGLFFKDTNSLAYYNVPPGAVIQLQIKERGGRKK
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
A0A2H1VZX9
A0A212EM12
H9JVN1
A0A194Q1L6
A0A0N1IFR8
A0A1B0CAM5
+ More
A0A1B0DEF2 A0A1L8DHC8 A0A0A9WXZ4 A0A0L0CQX4 A0A1B6CAA6 B0X8P5 A0A1B6H5H2 A0A084WCJ8 A0A2M4A443 A0A2M4BDT0 A0A1I8MY99 T1P7P7 A0A2M4BDS9 W5JSY5 B4NKJ9 A0A1I8Q357 Q297I7 B4KBM1 A0A2H8TVU8 A0A182MIT1 A0A0K8UKL3 A0A2S2NUU2 A0A0K8V196 A0A0A1WH51 W8C6H1 X1WRJ0 B4JHD7 A0A3B0K5A4 A0A1A9W587 A0A182Y6Z8 A0A182TYT8 A0A1Y9G8G6 A0A1B0AWJ9 A0A182RLJ9 A0A1A9Y5Y7 A0A158NLL3 A0A0K8UD64 A0A1A9Z7E6 A0A1Q3F5M0 A0A1A9V514 A0A2C9H8L5 A0A182LHA9 A0A182P2N1 A0A182H8J0 A0A1S4G009 A0A195B1P3 A0A0M8ZY16 Q16I26 A0A2A3ERQ6 A0A088AG06 A0A182Q9X0 A0A195F1X0 E9IJM9 F4WW66 A0A154NWS3 A0A182W1L7 A0A026WIG0 A0A182N3D1 A0A182HY57 A0A195CY75 A0A0L7QLN1 A0A151XCD9 A0A195E7C7 A0A0M4ENF6 J9JNE8 A0A310S558 E2C1B8 A0A2J7QUB2 K7ISL9 A0A067QI32 E2ATB4 A0A0J7KIW8 A0A182K806 A0A1B0FFR5 K7ISL2 A0A1J1J2X1 A0A0C9PSK9 A0A232EZM1
A0A1B0DEF2 A0A1L8DHC8 A0A0A9WXZ4 A0A0L0CQX4 A0A1B6CAA6 B0X8P5 A0A1B6H5H2 A0A084WCJ8 A0A2M4A443 A0A2M4BDT0 A0A1I8MY99 T1P7P7 A0A2M4BDS9 W5JSY5 B4NKJ9 A0A1I8Q357 Q297I7 B4KBM1 A0A2H8TVU8 A0A182MIT1 A0A0K8UKL3 A0A2S2NUU2 A0A0K8V196 A0A0A1WH51 W8C6H1 X1WRJ0 B4JHD7 A0A3B0K5A4 A0A1A9W587 A0A182Y6Z8 A0A182TYT8 A0A1Y9G8G6 A0A1B0AWJ9 A0A182RLJ9 A0A1A9Y5Y7 A0A158NLL3 A0A0K8UD64 A0A1A9Z7E6 A0A1Q3F5M0 A0A1A9V514 A0A2C9H8L5 A0A182LHA9 A0A182P2N1 A0A182H8J0 A0A1S4G009 A0A195B1P3 A0A0M8ZY16 Q16I26 A0A2A3ERQ6 A0A088AG06 A0A182Q9X0 A0A195F1X0 E9IJM9 F4WW66 A0A154NWS3 A0A182W1L7 A0A026WIG0 A0A182N3D1 A0A182HY57 A0A195CY75 A0A0L7QLN1 A0A151XCD9 A0A195E7C7 A0A0M4ENF6 J9JNE8 A0A310S558 E2C1B8 A0A2J7QUB2 K7ISL9 A0A067QI32 E2ATB4 A0A0J7KIW8 A0A182K806 A0A1B0FFR5 K7ISL2 A0A1J1J2X1 A0A0C9PSK9 A0A232EZM1
Pubmed
EMBL
ODYU01005188
SOQ45804.1
AGBW02013960
OWR42516.1
BABH01032330
KQ459579
+ More
KPI99472.1 KQ460650 KPJ13139.1 AJWK01004114 AJVK01032689 AJVK01032690 AJVK01032691 GFDF01008330 JAV05754.1 GBHO01030257 GDHC01001250 JAG13347.1 JAQ17379.1 JRES01000032 KNC34780.1 GEDC01026882 JAS10416.1 DS232495 EDS42637.1 GECU01037765 JAS69941.1 ATLV01022681 KE525335 KFB47942.1 GGFK01002252 MBW35573.1 GGFJ01002068 MBW51209.1 KA644504 AFP59133.1 GGFJ01002069 MBW51210.1 ADMH02000540 ETN66040.1 CH964272 EDW85171.2 CM000070 EAL28218.2 CH933806 EDW13688.1 GFXV01006589 MBW18394.1 AXCM01006132 GDHF01025231 GDHF01005979 JAI27083.1 JAI46335.1 GGMR01008278 MBY20897.1 GDHF01022549 GDHF01019688 JAI29765.1 JAI32626.1 GBXI01015948 GBXI01005256 JAC98343.1 JAD09036.1 GAMC01008541 JAB98014.1 ABLF02040727 CH916369 EDV93844.1 OUUW01000007 SPP83190.1 JXJN01004810 ADTU01019338 GDHF01028024 JAI24290.1 GFDL01012185 JAV22860.1 JXUM01118578 KQ566202 KXJ70321.1 KQ976662 KYM78413.1 KQ435801 KOX73257.1 CH478115 EAT33916.1 KZ288197 PBC33721.1 AXCN02001111 KQ981864 KYN34376.1 GL763877 EFZ19216.1 GL888404 EGI61537.1 KQ434773 KZC04012.1 KK107185 QOIP01000012 EZA55800.1 RLU15751.1 APCN01004493 KQ977115 KYN05620.1 KQ414914 KOC59537.1 KQ982314 KYQ58032.1 KQ979568 KYN20762.1 CP012526 ALC47140.1 ABLF02014752 KQ771002 OAD52554.1 GL451914 EFN78280.1 NEVH01011186 PNF32165.1 KK853363 KDR08142.1 GL442545 EFN63359.1 LBMM01006981 KMQ90146.1 CCAG010018629 CVRI01000064 CRL05201.1 GBYB01004323 GBYB01010738 JAG74090.1 JAG80505.1 NNAY01001464 OXU23884.1
KPI99472.1 KQ460650 KPJ13139.1 AJWK01004114 AJVK01032689 AJVK01032690 AJVK01032691 GFDF01008330 JAV05754.1 GBHO01030257 GDHC01001250 JAG13347.1 JAQ17379.1 JRES01000032 KNC34780.1 GEDC01026882 JAS10416.1 DS232495 EDS42637.1 GECU01037765 JAS69941.1 ATLV01022681 KE525335 KFB47942.1 GGFK01002252 MBW35573.1 GGFJ01002068 MBW51209.1 KA644504 AFP59133.1 GGFJ01002069 MBW51210.1 ADMH02000540 ETN66040.1 CH964272 EDW85171.2 CM000070 EAL28218.2 CH933806 EDW13688.1 GFXV01006589 MBW18394.1 AXCM01006132 GDHF01025231 GDHF01005979 JAI27083.1 JAI46335.1 GGMR01008278 MBY20897.1 GDHF01022549 GDHF01019688 JAI29765.1 JAI32626.1 GBXI01015948 GBXI01005256 JAC98343.1 JAD09036.1 GAMC01008541 JAB98014.1 ABLF02040727 CH916369 EDV93844.1 OUUW01000007 SPP83190.1 JXJN01004810 ADTU01019338 GDHF01028024 JAI24290.1 GFDL01012185 JAV22860.1 JXUM01118578 KQ566202 KXJ70321.1 KQ976662 KYM78413.1 KQ435801 KOX73257.1 CH478115 EAT33916.1 KZ288197 PBC33721.1 AXCN02001111 KQ981864 KYN34376.1 GL763877 EFZ19216.1 GL888404 EGI61537.1 KQ434773 KZC04012.1 KK107185 QOIP01000012 EZA55800.1 RLU15751.1 APCN01004493 KQ977115 KYN05620.1 KQ414914 KOC59537.1 KQ982314 KYQ58032.1 KQ979568 KYN20762.1 CP012526 ALC47140.1 ABLF02014752 KQ771002 OAD52554.1 GL451914 EFN78280.1 NEVH01011186 PNF32165.1 KK853363 KDR08142.1 GL442545 EFN63359.1 LBMM01006981 KMQ90146.1 CCAG010018629 CVRI01000064 CRL05201.1 GBYB01004323 GBYB01010738 JAG74090.1 JAG80505.1 NNAY01001464 OXU23884.1
Proteomes
UP000007151
UP000005204
UP000053268
UP000053240
UP000092461
UP000092462
+ More
UP000037069 UP000002320 UP000030765 UP000095301 UP000000673 UP000007798 UP000095300 UP000001819 UP000009192 UP000075883 UP000007819 UP000001070 UP000268350 UP000091820 UP000076408 UP000075902 UP000069272 UP000092460 UP000075900 UP000092443 UP000005205 UP000092445 UP000078200 UP000076407 UP000075882 UP000075885 UP000069940 UP000249989 UP000078540 UP000053105 UP000008820 UP000242457 UP000005203 UP000075886 UP000078541 UP000007755 UP000076502 UP000075920 UP000053097 UP000279307 UP000075884 UP000075840 UP000078542 UP000053825 UP000075809 UP000078492 UP000092553 UP000008237 UP000235965 UP000002358 UP000027135 UP000000311 UP000036403 UP000075881 UP000092444 UP000183832 UP000215335
UP000037069 UP000002320 UP000030765 UP000095301 UP000000673 UP000007798 UP000095300 UP000001819 UP000009192 UP000075883 UP000007819 UP000001070 UP000268350 UP000091820 UP000076408 UP000075902 UP000069272 UP000092460 UP000075900 UP000092443 UP000005205 UP000092445 UP000078200 UP000076407 UP000075882 UP000075885 UP000069940 UP000249989 UP000078540 UP000053105 UP000008820 UP000242457 UP000005203 UP000075886 UP000078541 UP000007755 UP000076502 UP000075920 UP000053097 UP000279307 UP000075884 UP000075840 UP000078542 UP000053825 UP000075809 UP000078492 UP000092553 UP000008237 UP000235965 UP000002358 UP000027135 UP000000311 UP000036403 UP000075881 UP000092444 UP000183832 UP000215335
PRIDE
Pfam
Interpro
IPR029071
Ubiquitin-like_domsf
+ More
IPR000061 Surp
IPR035967 SWAP/Surp_sf
IPR000626 Ubiquitin_dom
IPR022030 SF3A1
IPR008984 SMAD_FHA_dom_sf
IPR000253 FHA_dom
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR036961 Kinesin_motor_dom_sf
IPR035563 SF3As1_ubi
IPR019956 Ubiquitin
IPR029063 SAM-dependent_MTases
IPR007823 RRP8
IPR042036 RRP8_N
IPR000061 Surp
IPR035967 SWAP/Surp_sf
IPR000626 Ubiquitin_dom
IPR022030 SF3A1
IPR008984 SMAD_FHA_dom_sf
IPR000253 FHA_dom
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR036961 Kinesin_motor_dom_sf
IPR035563 SF3As1_ubi
IPR019956 Ubiquitin
IPR029063 SAM-dependent_MTases
IPR007823 RRP8
IPR042036 RRP8_N
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1VZX9
A0A212EM12
H9JVN1
A0A194Q1L6
A0A0N1IFR8
A0A1B0CAM5
+ More
A0A1B0DEF2 A0A1L8DHC8 A0A0A9WXZ4 A0A0L0CQX4 A0A1B6CAA6 B0X8P5 A0A1B6H5H2 A0A084WCJ8 A0A2M4A443 A0A2M4BDT0 A0A1I8MY99 T1P7P7 A0A2M4BDS9 W5JSY5 B4NKJ9 A0A1I8Q357 Q297I7 B4KBM1 A0A2H8TVU8 A0A182MIT1 A0A0K8UKL3 A0A2S2NUU2 A0A0K8V196 A0A0A1WH51 W8C6H1 X1WRJ0 B4JHD7 A0A3B0K5A4 A0A1A9W587 A0A182Y6Z8 A0A182TYT8 A0A1Y9G8G6 A0A1B0AWJ9 A0A182RLJ9 A0A1A9Y5Y7 A0A158NLL3 A0A0K8UD64 A0A1A9Z7E6 A0A1Q3F5M0 A0A1A9V514 A0A2C9H8L5 A0A182LHA9 A0A182P2N1 A0A182H8J0 A0A1S4G009 A0A195B1P3 A0A0M8ZY16 Q16I26 A0A2A3ERQ6 A0A088AG06 A0A182Q9X0 A0A195F1X0 E9IJM9 F4WW66 A0A154NWS3 A0A182W1L7 A0A026WIG0 A0A182N3D1 A0A182HY57 A0A195CY75 A0A0L7QLN1 A0A151XCD9 A0A195E7C7 A0A0M4ENF6 J9JNE8 A0A310S558 E2C1B8 A0A2J7QUB2 K7ISL9 A0A067QI32 E2ATB4 A0A0J7KIW8 A0A182K806 A0A1B0FFR5 K7ISL2 A0A1J1J2X1 A0A0C9PSK9 A0A232EZM1
A0A1B0DEF2 A0A1L8DHC8 A0A0A9WXZ4 A0A0L0CQX4 A0A1B6CAA6 B0X8P5 A0A1B6H5H2 A0A084WCJ8 A0A2M4A443 A0A2M4BDT0 A0A1I8MY99 T1P7P7 A0A2M4BDS9 W5JSY5 B4NKJ9 A0A1I8Q357 Q297I7 B4KBM1 A0A2H8TVU8 A0A182MIT1 A0A0K8UKL3 A0A2S2NUU2 A0A0K8V196 A0A0A1WH51 W8C6H1 X1WRJ0 B4JHD7 A0A3B0K5A4 A0A1A9W587 A0A182Y6Z8 A0A182TYT8 A0A1Y9G8G6 A0A1B0AWJ9 A0A182RLJ9 A0A1A9Y5Y7 A0A158NLL3 A0A0K8UD64 A0A1A9Z7E6 A0A1Q3F5M0 A0A1A9V514 A0A2C9H8L5 A0A182LHA9 A0A182P2N1 A0A182H8J0 A0A1S4G009 A0A195B1P3 A0A0M8ZY16 Q16I26 A0A2A3ERQ6 A0A088AG06 A0A182Q9X0 A0A195F1X0 E9IJM9 F4WW66 A0A154NWS3 A0A182W1L7 A0A026WIG0 A0A182N3D1 A0A182HY57 A0A195CY75 A0A0L7QLN1 A0A151XCD9 A0A195E7C7 A0A0M4ENF6 J9JNE8 A0A310S558 E2C1B8 A0A2J7QUB2 K7ISL9 A0A067QI32 E2ATB4 A0A0J7KIW8 A0A182K806 A0A1B0FFR5 K7ISL2 A0A1J1J2X1 A0A0C9PSK9 A0A232EZM1
PDB
6FF7
E-value=3.99116e-111,
Score=1029
Ontologies
GO
PANTHER
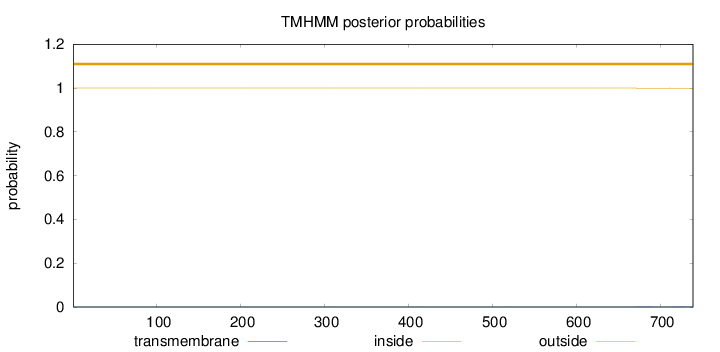
Topology
Length:
739
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01397
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 739
Population Genetic Test Statistics
Pi
248.92546
Theta
214.130714
Tajima's D
0.40209
CLR
0.228864
CSRT
0.482225888705565
Interpretation
Uncertain
