Gene
KWMTBOMO03244 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013593
Annotation
perilipin_[Bombyx_mori]
Full name
Lipid storage droplets surface-binding protein 1
Location in the cell
Mitochondrial Reliability : 2.356
Sequence
CDS
ATGAATGGTAATGGAGACGTTTCCAAAAAAAGCTTCAATGGCACATGTACGGTGGGCGTGAATAAAAAAGTCAAGGCTTCAATTGCTAGGAAAGCCGAGGCCTACCTGCAGGCTGTTAGTCAGAGTCAAGGATCTAAAGTGACGAAAACAAATCAGCCAGCCATGCCGAATCTTGAAGTGGTCTCAAGGGTTGCATCCATACCGATCGTCGTGTCCGGTATTGGAGTGACAGAAAAATTGTACTTCAAAATAAGGGAGTCCAACCCTCTGTTTCGTTGGTCTATGTCCCTGGGTGAGAAGTCCCTAGCGACCGGCATACAGCTAGCACTCCCGGCGGTTCAACTCCTGGAAACTCCGATTGTACAACTGGACAAGTTTCTCTGCAAGTCGCTGGATGTGGTCGAGAAGAGTATGCCTTCAATTTATATGCCTCCAGAGGAGATGTACTCAGAGACGCGTCAATACGTGCTCCGACGTGCGGATTCCGTGACGCGGCTAGGCACAGCTGTCTTGGATTCGCGGATGACGGCGGCCACCGCGACCGCTCTCGACCGTGCGCTCACCACCGCCGACAAATACGTCGACAAGTACCTCCCGCCAGACAACCAAGACGCAGCCGACGTGACCAGCGCTGATGCGGTGGACGGGCCTGAAGGGGAGACGGGCAGGGCCCGGGCAGCCCAGGCGGTGCAGCATGGAGCCAGGTTCAAGAGGAAGCTGCAGAGGAGACTCACCAGGCAAGCTCTGGCCGAAGCCAAGGCTATCAAAGAACAAATCCACGTTTTGGTGTATGTCGCGGAATTGGTTGCAAAAGATCCTGTGTTAGCCTGGAAAAAAGCTAAGGAACTTTACGCGTCTCTCAGTCAACCAGAGCCCGAGAATCAAGCGAGGCCTGTAACTCTTGAAGAGCTGATGGTACTCCTCACCAGGGAAACAGCAAGAAAGGTCGTGCATCTCGTCAACTATACACACACAGACCTTCCAAGAAACATCCGTCAAGGCATGTCGATTGTTACGAAGCATTTGTCGTATACAGCAGAGGCATTACTCAAGTCTGTACCCGTGGAGACGGCAATCACTGAAATAAAGGGATGGAGAAGCAAACTTGAGGTCCTGCTGCAGCAGTTGCAGGCCACATCCAAAACTTACTTGGAGCATTTAGCCATATTCTTGGCCGGTAACGAGGAGCGGGAGAAGATTGCGCCACGGTCGGCTTACGAGCAAAGAGACGACGTGTCTTCCATTAACGGTGTCAATTAA
Protein
MNGNGDVSKKSFNGTCTVGVNKKVKASIARKAEAYLQAVSQSQGSKVTKTNQPAMPNLEVVSRVASIPIVVSGIGVTEKLYFKIRESNPLFRWSMSLGEKSLATGIQLALPAVQLLETPIVQLDKFLCKSLDVVEKSMPSIYMPPEEMYSETRQYVLRRADSVTRLGTAVLDSRMTAATATALDRALTTADKYVDKYLPPDNQDAADVTSADAVDGPEGETGRARAAQAVQHGARFKRKLQRRLTRQALAEAKAIKEQIHVLVYVAELVAKDPVLAWKKAKELYASLSQPEPENQARPVTLEELMVLLTRETARKVVHLVNYTHTDLPRNIRQGMSIVTKHLSYTAEALLKSVPVETAITEIKGWRSKLEVLLQQLQATSKTYLEHLAIFLAGNEEREKIAPRSAYEQRDDVSSINGVN
Summary
Description
Required for normal deposition of neutral lipids in the oocyte.
Similarity
Belongs to the perilipin family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Lipid droplet
Lipid transport
Reference proteome
Transport
Feature
chain Lipid storage droplets surface-binding protein 1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JVN0
Q2F665
A0A068JFE1
A0A2W1BMU5
A0A194Q7T5
A0A068JIS1
+ More
B5A4A4 A0A0N1IPA5 A0A212EM14 D6WC49 V5I8M3 J3JZ70 A0A1I8N0R4 A0A1I8N0S4 A0A139WM71 B4K5B3 A0A1I8N0R7 A0A1I8PHE8 B4NG43 V5GIZ9 A0A336LF06 A0A0L0BUW8 B4JT97 B4R247 A0A0Q9XBZ6 A0A0C9RMP3 A0A0Q9WUF1 A0A088A728 A0A026WGN3 B4HFY5 B4M0F4 Q9VCI3-3 A0A0B4KHZ1 A0A1I8PHA8 A0A2A3E6E4 G0TEC0 A0A3B0KBJ8 B3LY50 V5G204 A0A0T6B2K4 Q299C6 A0A0A1WYW7 B3P8E2 A0A0M4F6S6 W8BBE6 E2BDR6 A0A291NUD1 B4PNX7 B4G510 A0A1W4UKA4 A0A034WM17 A0A0Q9XB11 D3TLD1 W8BZ49 A0A0P9AT90 A0A1A9X5Y7 A0A1B0ASN0 A0A336MG51 A0A0P6IW70 Q9VCI3 J3JUU0 E4NKM5 I0B1Q0 A0A1L8DZ84 A0A1W4UK68 A0A0L7QPV5 U4UPX1 A0A336MNM0 A0A1A9Z751 A0A1B0GIQ7 A0A0R1E8V9 A0A151WZK6 A0A1A9V5I1 A0A195EIZ7 F4WXT9 U5EJT6 A0A1I8PH32 A0A084VT42 A0A2M4AVW4 A0A2M4CPL9 A0A182K0S5 A0A1A9WVQ4 B0WA13 Q7QD01 A0A182J580 A0A195F5Z2 A0A182THY3 A0A2M3Z634 K7IWY0 A0A182HA85 A0A0C9R9N6 A0A2M3Z651 A0A2M3Z5U9 N6U0V8
B5A4A4 A0A0N1IPA5 A0A212EM14 D6WC49 V5I8M3 J3JZ70 A0A1I8N0R4 A0A1I8N0S4 A0A139WM71 B4K5B3 A0A1I8N0R7 A0A1I8PHE8 B4NG43 V5GIZ9 A0A336LF06 A0A0L0BUW8 B4JT97 B4R247 A0A0Q9XBZ6 A0A0C9RMP3 A0A0Q9WUF1 A0A088A728 A0A026WGN3 B4HFY5 B4M0F4 Q9VCI3-3 A0A0B4KHZ1 A0A1I8PHA8 A0A2A3E6E4 G0TEC0 A0A3B0KBJ8 B3LY50 V5G204 A0A0T6B2K4 Q299C6 A0A0A1WYW7 B3P8E2 A0A0M4F6S6 W8BBE6 E2BDR6 A0A291NUD1 B4PNX7 B4G510 A0A1W4UKA4 A0A034WM17 A0A0Q9XB11 D3TLD1 W8BZ49 A0A0P9AT90 A0A1A9X5Y7 A0A1B0ASN0 A0A336MG51 A0A0P6IW70 Q9VCI3 J3JUU0 E4NKM5 I0B1Q0 A0A1L8DZ84 A0A1W4UK68 A0A0L7QPV5 U4UPX1 A0A336MNM0 A0A1A9Z751 A0A1B0GIQ7 A0A0R1E8V9 A0A151WZK6 A0A1A9V5I1 A0A195EIZ7 F4WXT9 U5EJT6 A0A1I8PH32 A0A084VT42 A0A2M4AVW4 A0A2M4CPL9 A0A182K0S5 A0A1A9WVQ4 B0WA13 Q7QD01 A0A182J580 A0A195F5Z2 A0A182THY3 A0A2M3Z634 K7IWY0 A0A182HA85 A0A0C9R9N6 A0A2M3Z651 A0A2M3Z5U9 N6U0V8
Pubmed
19121390
28756777
26354079
15829485
18793726
22118469
+ More
18362917 19820115 22516182 25315136 17994087 26108605 24508170 30249741 12077142 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 21695433 15632085 25830018 24495485 20798317 17550304 25348373 20353571 26999592 23537049 21719571 24438588 12364791 14747013 17210077 20075255 26483478
18362917 19820115 22516182 25315136 17994087 26108605 24508170 30249741 12077142 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 21695433 15632085 25830018 24495485 20798317 17550304 25348373 20353571 26999592 23537049 21719571 24438588 12364791 14747013 17210077 20075255 26483478
EMBL
BABH01032332
BABH01032333
DQ311207
ABD36152.1
KF835604
AIE17454.1
+ More
KZ150071 PZC74040.1 KQ459579 KPI99470.1 KF835603 AIE17453.1 EU809925 ACF24761.1 KQ460650 KPJ13136.1 AGBW02013959 OWR42518.1 KQ971316 EEZ98760.2 GALX01004435 JAB64031.1 BT128551 AEE63508.1 KYB28986.1 CH933806 EDW16139.1 CH964251 EDW83260.1 GALX01004437 JAB64029.1 UFQS01001870 UFQT01001870 SSX12612.1 SSX32055.1 JRES01001300 KNC23788.1 CH916373 EDV94987.1 CM000364 EDX14104.1 KRG01953.1 GBYB01009635 JAG79402.1 CH940650 KRF83781.1 KK107218 QOIP01000001 EZA55190.1 RLU27651.1 CH480815 EDW43378.1 EDW68333.2 AF357214 AE014297 AY051436 AAF56183.2 AAK27222.1 AAK92860.1 AGB96263.1 KZ288353 PBC27285.1 GU722328 GU722332 ADT65132.1 OUUW01000007 SPP83046.1 CH902617 EDV43954.1 GALX01004436 JAB64030.1 LJIG01016077 KRT81683.1 CM000070 EAL27777.2 GBXI01010250 JAD04042.1 CH954182 EDV53966.1 CP012526 ALC47800.1 GAMC01007995 JAB98560.1 GL447689 EFN86122.1 MF124801 ATJ03455.1 CM000160 EDW98187.1 CH479179 EDW24676.1 GAKP01004134 JAC54818.1 KRG01954.1 EZ422233 ADD18509.1 GAMC01007994 JAB98561.1 KPU80560.1 JXJN01002890 UFQS01001180 UFQT01001180 SSX09553.1 SSX29354.1 GDUN01000543 JAN95376.1 BT127005 AEE61967.1 BT125822 ADR83707.1 BT133411 AFH55510.1 GFDF01002313 JAV11771.1 KQ414807 KOC60589.1 KB632314 ERL92191.1 SSX12611.1 SSX32054.1 AJWK01015297 AJWK01015298 AJWK01015299 KRK04145.1 KQ982649 KYQ53197.1 KQ978801 KYN28235.1 GL888427 EGI61017.1 GANO01002105 JAB57766.1 ATLV01016166 ATLV01016167 KE525060 KFB41136.1 GGFK01011447 MBW44768.1 GGFL01003065 MBW67243.1 DS231868 EDS40746.1 AAAB01008859 EAA07592.5 KQ981805 KYN35489.1 GGFM01003219 MBW23970.1 AAZX01004124 JXUM01030863 KQ560899 KXJ80427.1 GBYB01009637 JAG79404.1 GGFM01003224 MBW23975.1 GGFM01003156 MBW23907.1 APGK01053890 APGK01053891 APGK01053892 APGK01053893 APGK01053894 APGK01053895 APGK01053896 APGK01053897 APGK01053898 APGK01053899 KB741238 ENN72182.1
KZ150071 PZC74040.1 KQ459579 KPI99470.1 KF835603 AIE17453.1 EU809925 ACF24761.1 KQ460650 KPJ13136.1 AGBW02013959 OWR42518.1 KQ971316 EEZ98760.2 GALX01004435 JAB64031.1 BT128551 AEE63508.1 KYB28986.1 CH933806 EDW16139.1 CH964251 EDW83260.1 GALX01004437 JAB64029.1 UFQS01001870 UFQT01001870 SSX12612.1 SSX32055.1 JRES01001300 KNC23788.1 CH916373 EDV94987.1 CM000364 EDX14104.1 KRG01953.1 GBYB01009635 JAG79402.1 CH940650 KRF83781.1 KK107218 QOIP01000001 EZA55190.1 RLU27651.1 CH480815 EDW43378.1 EDW68333.2 AF357214 AE014297 AY051436 AAF56183.2 AAK27222.1 AAK92860.1 AGB96263.1 KZ288353 PBC27285.1 GU722328 GU722332 ADT65132.1 OUUW01000007 SPP83046.1 CH902617 EDV43954.1 GALX01004436 JAB64030.1 LJIG01016077 KRT81683.1 CM000070 EAL27777.2 GBXI01010250 JAD04042.1 CH954182 EDV53966.1 CP012526 ALC47800.1 GAMC01007995 JAB98560.1 GL447689 EFN86122.1 MF124801 ATJ03455.1 CM000160 EDW98187.1 CH479179 EDW24676.1 GAKP01004134 JAC54818.1 KRG01954.1 EZ422233 ADD18509.1 GAMC01007994 JAB98561.1 KPU80560.1 JXJN01002890 UFQS01001180 UFQT01001180 SSX09553.1 SSX29354.1 GDUN01000543 JAN95376.1 BT127005 AEE61967.1 BT125822 ADR83707.1 BT133411 AFH55510.1 GFDF01002313 JAV11771.1 KQ414807 KOC60589.1 KB632314 ERL92191.1 SSX12611.1 SSX32054.1 AJWK01015297 AJWK01015298 AJWK01015299 KRK04145.1 KQ982649 KYQ53197.1 KQ978801 KYN28235.1 GL888427 EGI61017.1 GANO01002105 JAB57766.1 ATLV01016166 ATLV01016167 KE525060 KFB41136.1 GGFK01011447 MBW44768.1 GGFL01003065 MBW67243.1 DS231868 EDS40746.1 AAAB01008859 EAA07592.5 KQ981805 KYN35489.1 GGFM01003219 MBW23970.1 AAZX01004124 JXUM01030863 KQ560899 KXJ80427.1 GBYB01009637 JAG79404.1 GGFM01003224 MBW23975.1 GGFM01003156 MBW23907.1 APGK01053890 APGK01053891 APGK01053892 APGK01053893 APGK01053894 APGK01053895 APGK01053896 APGK01053897 APGK01053898 APGK01053899 KB741238 ENN72182.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000007266
UP000095301
+ More
UP000009192 UP000095300 UP000007798 UP000037069 UP000001070 UP000000304 UP000008792 UP000005203 UP000053097 UP000279307 UP000001292 UP000000803 UP000242457 UP000268350 UP000007801 UP000001819 UP000008711 UP000092553 UP000008237 UP000002282 UP000008744 UP000192221 UP000092443 UP000092460 UP000053825 UP000030742 UP000092445 UP000092461 UP000075809 UP000078200 UP000078492 UP000007755 UP000030765 UP000075881 UP000091820 UP000002320 UP000007062 UP000075880 UP000078541 UP000075902 UP000002358 UP000069940 UP000249989 UP000019118
UP000009192 UP000095300 UP000007798 UP000037069 UP000001070 UP000000304 UP000008792 UP000005203 UP000053097 UP000279307 UP000001292 UP000000803 UP000242457 UP000268350 UP000007801 UP000001819 UP000008711 UP000092553 UP000008237 UP000002282 UP000008744 UP000192221 UP000092443 UP000092460 UP000053825 UP000030742 UP000092445 UP000092461 UP000075809 UP000078200 UP000078492 UP000007755 UP000030765 UP000075881 UP000091820 UP000002320 UP000007062 UP000075880 UP000078541 UP000075902 UP000002358 UP000069940 UP000249989 UP000019118
Pfam
PF03036 Perilipin
CDD
ProteinModelPortal
H9JVN0
Q2F665
A0A068JFE1
A0A2W1BMU5
A0A194Q7T5
A0A068JIS1
+ More
B5A4A4 A0A0N1IPA5 A0A212EM14 D6WC49 V5I8M3 J3JZ70 A0A1I8N0R4 A0A1I8N0S4 A0A139WM71 B4K5B3 A0A1I8N0R7 A0A1I8PHE8 B4NG43 V5GIZ9 A0A336LF06 A0A0L0BUW8 B4JT97 B4R247 A0A0Q9XBZ6 A0A0C9RMP3 A0A0Q9WUF1 A0A088A728 A0A026WGN3 B4HFY5 B4M0F4 Q9VCI3-3 A0A0B4KHZ1 A0A1I8PHA8 A0A2A3E6E4 G0TEC0 A0A3B0KBJ8 B3LY50 V5G204 A0A0T6B2K4 Q299C6 A0A0A1WYW7 B3P8E2 A0A0M4F6S6 W8BBE6 E2BDR6 A0A291NUD1 B4PNX7 B4G510 A0A1W4UKA4 A0A034WM17 A0A0Q9XB11 D3TLD1 W8BZ49 A0A0P9AT90 A0A1A9X5Y7 A0A1B0ASN0 A0A336MG51 A0A0P6IW70 Q9VCI3 J3JUU0 E4NKM5 I0B1Q0 A0A1L8DZ84 A0A1W4UK68 A0A0L7QPV5 U4UPX1 A0A336MNM0 A0A1A9Z751 A0A1B0GIQ7 A0A0R1E8V9 A0A151WZK6 A0A1A9V5I1 A0A195EIZ7 F4WXT9 U5EJT6 A0A1I8PH32 A0A084VT42 A0A2M4AVW4 A0A2M4CPL9 A0A182K0S5 A0A1A9WVQ4 B0WA13 Q7QD01 A0A182J580 A0A195F5Z2 A0A182THY3 A0A2M3Z634 K7IWY0 A0A182HA85 A0A0C9R9N6 A0A2M3Z651 A0A2M3Z5U9 N6U0V8
B5A4A4 A0A0N1IPA5 A0A212EM14 D6WC49 V5I8M3 J3JZ70 A0A1I8N0R4 A0A1I8N0S4 A0A139WM71 B4K5B3 A0A1I8N0R7 A0A1I8PHE8 B4NG43 V5GIZ9 A0A336LF06 A0A0L0BUW8 B4JT97 B4R247 A0A0Q9XBZ6 A0A0C9RMP3 A0A0Q9WUF1 A0A088A728 A0A026WGN3 B4HFY5 B4M0F4 Q9VCI3-3 A0A0B4KHZ1 A0A1I8PHA8 A0A2A3E6E4 G0TEC0 A0A3B0KBJ8 B3LY50 V5G204 A0A0T6B2K4 Q299C6 A0A0A1WYW7 B3P8E2 A0A0M4F6S6 W8BBE6 E2BDR6 A0A291NUD1 B4PNX7 B4G510 A0A1W4UKA4 A0A034WM17 A0A0Q9XB11 D3TLD1 W8BZ49 A0A0P9AT90 A0A1A9X5Y7 A0A1B0ASN0 A0A336MG51 A0A0P6IW70 Q9VCI3 J3JUU0 E4NKM5 I0B1Q0 A0A1L8DZ84 A0A1W4UK68 A0A0L7QPV5 U4UPX1 A0A336MNM0 A0A1A9Z751 A0A1B0GIQ7 A0A0R1E8V9 A0A151WZK6 A0A1A9V5I1 A0A195EIZ7 F4WXT9 U5EJT6 A0A1I8PH32 A0A084VT42 A0A2M4AVW4 A0A2M4CPL9 A0A182K0S5 A0A1A9WVQ4 B0WA13 Q7QD01 A0A182J580 A0A195F5Z2 A0A182THY3 A0A2M3Z634 K7IWY0 A0A182HA85 A0A0C9R9N6 A0A2M3Z651 A0A2M3Z5U9 N6U0V8
PDB
2DWQ
E-value=0.00788981,
Score=93
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Lipid droplet
Lipid droplet
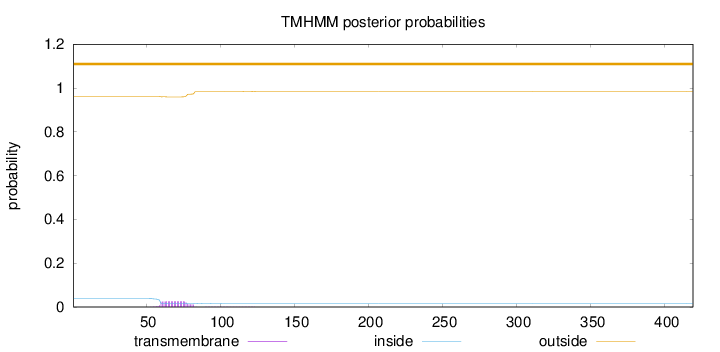
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.550859999999999
Exp number, first 60 AAs:
0.04303
Total prob of N-in:
0.03857
outside
1 - 419
Population Genetic Test Statistics
Pi
21.602384
Theta
160.506677
Tajima's D
0.517904
CLR
0.099297
CSRT
0.522673866306685
Interpretation
Uncertain
