Pre Gene Modal
BGIBMGA013624
Annotation
PREDICTED:_suppressor_of_SWI4_1_homolog_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 4.187
Sequence
CDS
ATGGGGAAACGTAAAGGAAAATGCGTGAAGAAGAATTCCGCAAGTAAAGAAGCTTTGGAGCCTGAAAACCTAGTGAAAGCGCCACATTCGTTCGTGATTCACAGAGGGCAATGTAGCAAGGACCTCGTAGACTTAACTAGAGACTTCAGAAAGATCATGGAACCTTTCACTGCTTCACAATTAAAAGAACGCAAGAAAAATACAATTAAAGATTTTGTGTCCATATCTGGTTATCTTCATGTATCACACATGGTCCTTTTTACGGAAACTGAACTTGGTTCTTACATGAGACTTGCCAGACTACCTCGAGGGCCTACACTAACTTTTAAAATACACAATTACAGTTTAGCTCGTGATGTAATATCTTCACTGAAAAAACAATATGTGATGATGCGTGCCTTCCAAAATGCACCTCTCATTGTGCTAAACACCTTCTCTGGTGAAGGCATGCACATGAAACTTATGGCCACAATGTTTCAAAATATGTTTCCAACAATTAATATTACTACTGTAAAACTAAAAAATATAAGGAGATGTGTATTAATGAACTACAATCCGTCCACAAAACTAATAGATATGAGGCATTATGTAATTAGGGCAACACCGGTTGGACTTAATAAAGGAACTAAAAAGGTGGTCCAAGGCAAAATACCAAACTTGAACCGATGTAAAGATATGTCCGAGTTCTTTGACAAAGCGGGCTTGCTATCTGAGAGTGAGTTCGAGGATGATCCAAATTCTCAAATAGTTCTTCCACAAAGTCTGGCGTCGAGGGGAGCAGCCGTTGATTCTAAATCAGCTATCAGGTTATTTGAGCTCGGACCAAGGATAACATTTCAACTTATAAAGGTTGAAGATGGTCTCATGGATGGTGAAGTTTTATACCATGAGCTTGTAGAAAAAACAGAAGAAGAAAAAGCATTAATTAAAAAGAGAAGAGAGGCGAAGAGACGTCTAAAAGAAAAACGTAAATCGAAACAAGAAGAGAACATTAAGAAGAAGCAGAAAGAAAAAGAAGAACTCAAACAGAAGTCTCTTGAAGGCATTAAGAAGAAGAAAGAATCCACTGAGAGTCAGAGGTTAATGGAACTGGCGGCCGCCGAGTCTCAGAACAATGATCAAGATATGGAGGAAGACGACGACGCTGATTACTACAGACAGGAAGTGGGAGCCGAACCGGAAAAAGATCTGTTCACACCCAACACGTCGAAAAAGAGAAAGGCTGATGACAATTCTGTAAGAGGTTTCAAAAAAATGCGGCTCGACAAGAAATTAAAAAGGAAATATCAGAAAAATCAAGATGGCGGCCAACAGACAAAGAAACAAGGCAACGACAAGAAGTTCCAAAATAAGGATTTCAAAAATAAGAATTCAGGAGATAGAGATAAATTTAGGAATAAAGGTCAGAAGAAAGTGTTTGGAGGGAAAGTTCAGAATAAGTTTAATAAAGGCAGCAAAGGGAGGGAAGGGAAAGGTAGAAAGAACTCCGGCAAAGGAAAAGGGCGGAAATAA
Protein
MGKRKGKCVKKNSASKEALEPENLVKAPHSFVIHRGQCSKDLVDLTRDFRKIMEPFTASQLKERKKNTIKDFVSISGYLHVSHMVLFTETELGSYMRLARLPRGPTLTFKIHNYSLARDVISSLKKQYVMMRAFQNAPLIVLNTFSGEGMHMKLMATMFQNMFPTINITTVKLKNIRRCVLMNYNPSTKLIDMRHYVIRATPVGLNKGTKKVVQGKIPNLNRCKDMSEFFDKAGLLSESEFEDDPNSQIVLPQSLASRGAAVDSKSAIRLFELGPRITFQLIKVEDGLMDGEVLYHELVEKTEEEKALIKKRREAKRRLKEKRKSKQEENIKKKQKEKEELKQKSLEGIKKKKESTESQRLMELAAAESQNNDQDMEEDDDADYYRQEVGAEPEKDLFTPNTSKKRKADDNSVRGFKKMRLDKKLKRKYQKNQDGGQQTKKQGNDKKFQNKDFKNKNSGDRDKFRNKGQKKVFGGKVQNKFNKGSKGREGKGRKNSGKGKGRK
Summary
Uniprot
A0A2W1BG27
A0A2A4J5A1
A0A212FKA3
A0A2H1WL87
A0A0N1IHI2
S4NWG0
+ More
D6W8M2 A0A2J7QHE9 A0A232F2I0 F4WWW6 A0A195BY66 A0A158NCC8 A0A026WV10 A0A067RCQ8 A0A151IG90 K7IZ67 A0A0J7L325 A0A151WFU5 E2B1W3 A0A2P8XET7 A0A0T6B8K1 A0A088AIC5 E9J841 A0A310SMT2 A0A0L7RDS8 A0A1B6GAG7 A0A1B6KUH2 A0A154P4Y9 A0A1B6IDJ2 A0A0M9A7A9 A0A069DU24 A0A1S3CZV3 R4G3J6 A0A224XPA2 A0A2J7QHD2 A0A0K8THP6 A0A146M915 A0A0A9WJU8 A0A2R5L656 E2BI79 A0A151J5H8 A0A195FAK5 A0A2R7VWI6 A0A0A1WQJ4 A0A034WB35 A0A0M5J1U6 A0A1I8P0S5 B4JFI7 A0A0P5DHR0 A0A0P6IM96 A0A0P4YBK0 E9HQF0 A0A0P4Y3B3 B4LWU8
D6W8M2 A0A2J7QHE9 A0A232F2I0 F4WWW6 A0A195BY66 A0A158NCC8 A0A026WV10 A0A067RCQ8 A0A151IG90 K7IZ67 A0A0J7L325 A0A151WFU5 E2B1W3 A0A2P8XET7 A0A0T6B8K1 A0A088AIC5 E9J841 A0A310SMT2 A0A0L7RDS8 A0A1B6GAG7 A0A1B6KUH2 A0A154P4Y9 A0A1B6IDJ2 A0A0M9A7A9 A0A069DU24 A0A1S3CZV3 R4G3J6 A0A224XPA2 A0A2J7QHD2 A0A0K8THP6 A0A146M915 A0A0A9WJU8 A0A2R5L656 E2BI79 A0A151J5H8 A0A195FAK5 A0A2R7VWI6 A0A0A1WQJ4 A0A034WB35 A0A0M5J1U6 A0A1I8P0S5 B4JFI7 A0A0P5DHR0 A0A0P6IM96 A0A0P4YBK0 E9HQF0 A0A0P4Y3B3 B4LWU8
Pubmed
EMBL
KZ150071
PZC74032.1
NWSH01003045
PCG67059.1
AGBW02008137
OWR54129.1
+ More
ODYU01009332 SOQ53682.1 KQ460542 KPJ14115.1 GAIX01014675 JAA77885.1 KQ971312 EEZ98288.1 NEVH01013976 PNF27996.1 NNAY01001222 OXU24688.1 GL888417 EGI61209.1 KQ976396 KYM92883.1 ADTU01011814 KK107087 QOIP01000001 EZA59852.1 RLU27436.1 KK852785 KDR16561.1 KQ977728 KYN00255.1 LBMM01000915 KMQ97227.1 KQ983211 KYQ46665.1 GL444981 EFN60319.1 PYGN01002421 PSN30520.1 LJIG01009146 KRT83663.1 GL768788 EFZ11018.1 KQ760869 OAD58798.1 KQ414614 KOC68886.1 GECZ01010343 JAS59426.1 GEBQ01024887 JAT15090.1 KQ434820 KZC06996.1 GECU01022712 JAS84994.1 KQ435720 KOX78687.1 GBGD01001276 JAC87613.1 ACPB03009759 GAHY01001621 JAA75889.1 GFTR01006597 JAW09829.1 PNF27995.1 GBRD01000713 JAG65108.1 GDHC01002385 JAQ16244.1 GBHO01034877 JAG08727.1 GGLE01000791 MBY04917.1 GL448457 EFN84610.1 KQ979973 KYN18371.1 KQ981727 KYN37074.1 KK854049 PTY10285.1 GBXI01013150 JAD01142.1 GAKP01007612 JAC51340.1 CP012526 ALC45761.1 CH916369 EDV93468.1 GDIP01157336 JAJ66066.1 GDIQ01002835 LRGB01000512 JAN91902.1 KZS18504.1 GDIP01230787 JAI92614.1 GL732719 EFX66032.1 GDIP01233151 JAI90250.1 CH940650 EDW66669.1
ODYU01009332 SOQ53682.1 KQ460542 KPJ14115.1 GAIX01014675 JAA77885.1 KQ971312 EEZ98288.1 NEVH01013976 PNF27996.1 NNAY01001222 OXU24688.1 GL888417 EGI61209.1 KQ976396 KYM92883.1 ADTU01011814 KK107087 QOIP01000001 EZA59852.1 RLU27436.1 KK852785 KDR16561.1 KQ977728 KYN00255.1 LBMM01000915 KMQ97227.1 KQ983211 KYQ46665.1 GL444981 EFN60319.1 PYGN01002421 PSN30520.1 LJIG01009146 KRT83663.1 GL768788 EFZ11018.1 KQ760869 OAD58798.1 KQ414614 KOC68886.1 GECZ01010343 JAS59426.1 GEBQ01024887 JAT15090.1 KQ434820 KZC06996.1 GECU01022712 JAS84994.1 KQ435720 KOX78687.1 GBGD01001276 JAC87613.1 ACPB03009759 GAHY01001621 JAA75889.1 GFTR01006597 JAW09829.1 PNF27995.1 GBRD01000713 JAG65108.1 GDHC01002385 JAQ16244.1 GBHO01034877 JAG08727.1 GGLE01000791 MBY04917.1 GL448457 EFN84610.1 KQ979973 KYN18371.1 KQ981727 KYN37074.1 KK854049 PTY10285.1 GBXI01013150 JAD01142.1 GAKP01007612 JAC51340.1 CP012526 ALC45761.1 CH916369 EDV93468.1 GDIP01157336 JAJ66066.1 GDIQ01002835 LRGB01000512 JAN91902.1 KZS18504.1 GDIP01230787 JAI92614.1 GL732719 EFX66032.1 GDIP01233151 JAI90250.1 CH940650 EDW66669.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000007266
UP000235965
UP000215335
+ More
UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000027135 UP000078542 UP000002358 UP000036403 UP000075809 UP000000311 UP000245037 UP000005203 UP000053825 UP000076502 UP000053105 UP000079169 UP000015103 UP000008237 UP000078492 UP000078541 UP000092553 UP000095300 UP000001070 UP000076858 UP000000305 UP000008792
UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000027135 UP000078542 UP000002358 UP000036403 UP000075809 UP000000311 UP000245037 UP000005203 UP000053825 UP000076502 UP000053105 UP000079169 UP000015103 UP000008237 UP000078492 UP000078541 UP000092553 UP000095300 UP000001070 UP000076858 UP000000305 UP000008792
Pfam
PF04427 Brix
Interpro
IPR007109
Brix
ProteinModelPortal
A0A2W1BG27
A0A2A4J5A1
A0A212FKA3
A0A2H1WL87
A0A0N1IHI2
S4NWG0
+ More
D6W8M2 A0A2J7QHE9 A0A232F2I0 F4WWW6 A0A195BY66 A0A158NCC8 A0A026WV10 A0A067RCQ8 A0A151IG90 K7IZ67 A0A0J7L325 A0A151WFU5 E2B1W3 A0A2P8XET7 A0A0T6B8K1 A0A088AIC5 E9J841 A0A310SMT2 A0A0L7RDS8 A0A1B6GAG7 A0A1B6KUH2 A0A154P4Y9 A0A1B6IDJ2 A0A0M9A7A9 A0A069DU24 A0A1S3CZV3 R4G3J6 A0A224XPA2 A0A2J7QHD2 A0A0K8THP6 A0A146M915 A0A0A9WJU8 A0A2R5L656 E2BI79 A0A151J5H8 A0A195FAK5 A0A2R7VWI6 A0A0A1WQJ4 A0A034WB35 A0A0M5J1U6 A0A1I8P0S5 B4JFI7 A0A0P5DHR0 A0A0P6IM96 A0A0P4YBK0 E9HQF0 A0A0P4Y3B3 B4LWU8
D6W8M2 A0A2J7QHE9 A0A232F2I0 F4WWW6 A0A195BY66 A0A158NCC8 A0A026WV10 A0A067RCQ8 A0A151IG90 K7IZ67 A0A0J7L325 A0A151WFU5 E2B1W3 A0A2P8XET7 A0A0T6B8K1 A0A088AIC5 E9J841 A0A310SMT2 A0A0L7RDS8 A0A1B6GAG7 A0A1B6KUH2 A0A154P4Y9 A0A1B6IDJ2 A0A0M9A7A9 A0A069DU24 A0A1S3CZV3 R4G3J6 A0A224XPA2 A0A2J7QHD2 A0A0K8THP6 A0A146M915 A0A0A9WJU8 A0A2R5L656 E2BI79 A0A151J5H8 A0A195FAK5 A0A2R7VWI6 A0A0A1WQJ4 A0A034WB35 A0A0M5J1U6 A0A1I8P0S5 B4JFI7 A0A0P5DHR0 A0A0P6IM96 A0A0P4YBK0 E9HQF0 A0A0P4Y3B3 B4LWU8
PDB
6C0F
E-value=1.09593e-41,
Score=429
Ontologies
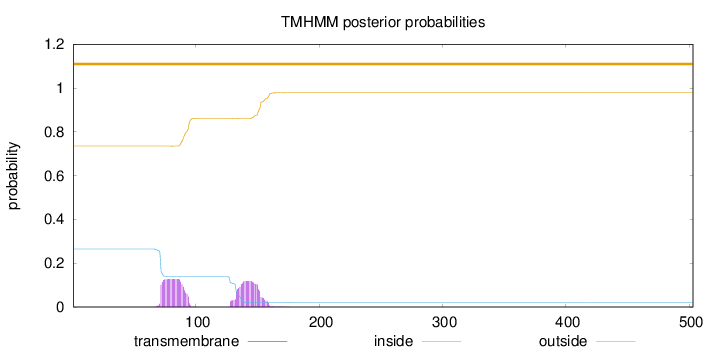
Topology
Length:
503
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.09002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.26498
outside
1 - 503
Population Genetic Test Statistics
Pi
237.538019
Theta
178.816507
Tajima's D
0.870026
CLR
0.034101
CSRT
0.623468826558672
Interpretation
Uncertain
