Pre Gene Modal
BGIBMGA013589
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_ER_membrane_protein_complex_subunit_2-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.26
Sequence
CDS
ATGTCTTTTAATTACGAAGAAATAAGCCACAATGAAGCAAAGGATCTTCTCCGTCAATGGAGGGAAAACAATGAGAGGAGAGGCAAAGAAGTGCTAGAACTTTGGGATGTGGTTCTCAGAGAAGATGTAAACAAACTTGGAAACGAGAAACACGTTATATTGGAGCAAGTAATCTATGCAGCATTGGACTGTCACATGTACTGCATAGCAATGTTGTGCCTTGTGATGCTATCCAATGAGTTTCCTGGTAGCCTAAGAGTTATGAAATTAAAAGCAGCTGTTTTAGAGGCAGAGGAGAAGTTCGATGAAGCTTTAGAATTACTGGACAACATAATCAAAGTAGATGAAACTAATTCAGCAGCACGGAAGCGCCGGATCGCGATTCTGAAAGCTCAAGGTTACATAGTCGAAGCGATCAAGGAACTTGTGGACTATTTGAAAAAGTTTATGTCAGATGTGGAAGCCTGGCAAGAGCTTTGCAACCTGTACCTTCAGGTTCAGGACTACTCTCGTGCCGTGTTCTGCGCCGAAGAACTACTCCTCCATCAGCCTCACAGCCACTTGCATCATCAGAGGCTAGCTGATATTCGGTACACAATGGGCGGCATCGAAAATCTGGAGTTGGCAAAACTGTATTACTGTCAATCTGTGAAACTCAATCCAGATAATATGAGGTCGCTGCTGGGTCTGTACTTGACAACAACAAATCTACTGAACCACTACAAATCATCAGGGAACAGTGGCAAGCGCAAGGAAGTGTGGAAACTGTGCCACTGGGCTCAGTCCAAAGCAACCAAACAAACTCAGCCGACAAGTCAACTGTCCCAGATGATGCTATCGTTGGCCATAACTGAATAA
Protein
MSFNYEEISHNEAKDLLRQWRENNERRGKEVLELWDVVLREDVNKLGNEKHVILEQVIYAALDCHMYCIAMLCLVMLSNEFPGSLRVMKLKAAVLEAEEKFDEALELLDNIIKVDETNSAARKRRIAILKAQGYIVEAIKELVDYLKKFMSDVEAWQELCNLYLQVQDYSRAVFCAEELLLHQPHSHLHHQRLADIRYTMGGIENLELAKLYYCQSVKLNPDNMRSLLGLYLTTTNLLNHYKSSGNSGKRKEVWKLCHWAQSKATKQTQPTSQLSQMMLSLAITE
Summary
Uniprot
H9JVM6
A0A2W1BGS9
A0A2H1WS25
I4DKH2
I4DS74
A0A0N0PCX0
+ More
S4NQJ4 A0A1E1WCU3 A0A212F6U5 A0A194Q1N1 A0A2A4J4Z3 A0A182XNZ1 A0A1I8Q7H9 A0A0L0CTR6 A0A182VEK3 A0A182TIY1 A0A182HK37 A0A1B0CR55 Q7PWZ6 A0A1I8MPL5 A0A2Y9D1G2 A0A0K8TQ66 A0A1L8E672 A0A1A9Y5Z0 A0A182JRG8 A0A182QE33 A0A182NR02 B4M5K3 A0A1B0AWJ2 A0A1A9ZFL9 A0A1A9UNW5 B3M2C0 D3TMX9 A0A1A9W598 A0A336M1A8 A0A182MGA1 A0A182YNB7 A0A182VW81 A0A336MI30 A0A1Q3F9Y8 A0A1B0D783 W5J5B0 A0A026X0R4 B4KBL9 A0A2A3EGP4 V9IH92 A0A088ATH4 A0A3B0JLP8 Q297I9 B4G4J7 A0A182FRZ4 A0A0T6AW84 A0A182PR69 A0A182RG10 W8CDS6 B0WYS9 A0A2M3ZI91 A0A182SAG7 F4X127 A0A195DWZ7 A0A0M4ERQ5 A0A195FQ25 A0A151XDE2 A0A023EP50 B4JHD5 Q16NY9 A0A195B6K9 A0A158NGY3 A0A1J1HKD6 A0A154NX87 A0A2M4BUW3 Q1HQY8 A0A0L7R806 A0A1W4VI77 U5ERN5 E2AII0 A0A1B0FFQ6 Q8SXY3 A0A2M4ALP4 A0A0A1XEQ9 A0A084W071 B3NZA5 Q9VEQ1 B4PM99 Q9VEQ2 Q8SZM8 B3NZA7 A0A182HAB0 B4QWE1 A0A034W8T7 B4QWD9 K7J067 A0A3B0KBX1 B4NKJ7 A0A1Y1KP61 A0A310S720 A0A0N7Z965 B4IBP3 R4FQM4 D6WAQ9
S4NQJ4 A0A1E1WCU3 A0A212F6U5 A0A194Q1N1 A0A2A4J4Z3 A0A182XNZ1 A0A1I8Q7H9 A0A0L0CTR6 A0A182VEK3 A0A182TIY1 A0A182HK37 A0A1B0CR55 Q7PWZ6 A0A1I8MPL5 A0A2Y9D1G2 A0A0K8TQ66 A0A1L8E672 A0A1A9Y5Z0 A0A182JRG8 A0A182QE33 A0A182NR02 B4M5K3 A0A1B0AWJ2 A0A1A9ZFL9 A0A1A9UNW5 B3M2C0 D3TMX9 A0A1A9W598 A0A336M1A8 A0A182MGA1 A0A182YNB7 A0A182VW81 A0A336MI30 A0A1Q3F9Y8 A0A1B0D783 W5J5B0 A0A026X0R4 B4KBL9 A0A2A3EGP4 V9IH92 A0A088ATH4 A0A3B0JLP8 Q297I9 B4G4J7 A0A182FRZ4 A0A0T6AW84 A0A182PR69 A0A182RG10 W8CDS6 B0WYS9 A0A2M3ZI91 A0A182SAG7 F4X127 A0A195DWZ7 A0A0M4ERQ5 A0A195FQ25 A0A151XDE2 A0A023EP50 B4JHD5 Q16NY9 A0A195B6K9 A0A158NGY3 A0A1J1HKD6 A0A154NX87 A0A2M4BUW3 Q1HQY8 A0A0L7R806 A0A1W4VI77 U5ERN5 E2AII0 A0A1B0FFQ6 Q8SXY3 A0A2M4ALP4 A0A0A1XEQ9 A0A084W071 B3NZA5 Q9VEQ1 B4PM99 Q9VEQ2 Q8SZM8 B3NZA7 A0A182HAB0 B4QWE1 A0A034W8T7 B4QWD9 K7J067 A0A3B0KBX1 B4NKJ7 A0A1Y1KP61 A0A310S720 A0A0N7Z965 B4IBP3 R4FQM4 D6WAQ9
Pubmed
19121390
28756777
22651552
26354079
23622113
22118469
+ More
26108605 12364791 14747013 17210077 25315136 20966253 26369729 17994087 20353571 25244985 20920257 23761445 24508170 30249741 15632085 24495485 21719571 24945155 17510324 21347285 17204158 20798317 25830018 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26483478 25348373 20075255 28004739 27129103 18362917 19820115
26108605 12364791 14747013 17210077 25315136 20966253 26369729 17994087 20353571 25244985 20920257 23761445 24508170 30249741 15632085 24495485 21719571 24945155 17510324 21347285 17204158 20798317 25830018 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26483478 25348373 20075255 28004739 27129103 18362917 19820115
EMBL
BABH01032365
KZ150071
PZC74028.1
ODYU01010626
SOQ55863.1
AK401790
+ More
BAM18412.1 AK405452 BAM20764.1 KQ460542 KPJ14122.1 GAIX01011519 JAA81041.1 GDQN01006373 JAT84681.1 AGBW02009968 OWR49461.1 KQ459579 KPI99487.1 NWSH01003045 PCG67051.1 JRES01000032 KNC34779.1 APCN01000816 AJWK01024414 AAAB01008987 EAA00991.4 GDAI01001568 JAI16035.1 GFDF01000039 JAV14045.1 AXCN02001485 CH940652 EDW58929.1 JXJN01004809 CH902617 EDV42311.1 EZ422781 ADD19057.1 UFQT01000415 SSX24106.1 AXCM01000086 UFQS01001255 UFQT01001255 SSX09952.1 SSX29675.1 GFDL01010679 JAV24366.1 AJVK01026700 ADMH02002101 ETN59171.1 KK107063 QOIP01000005 EZA60984.1 RLU23189.1 CH933806 EDW13686.1 KZ288253 PBC30935.1 JR047461 AEY60425.1 OUUW01000007 SPP83187.1 CM000070 EAL28216.1 CH479179 EDW25168.1 LJIG01022719 KRT79070.1 GAMC01001431 JAC05125.1 DS232197 EDS37189.1 GGFM01007409 MBW28160.1 GL888525 EGI59838.1 KQ980204 KYN17242.1 CP012526 ALC47888.1 KQ981387 KYN42024.1 KQ982294 KYQ58288.1 GAPW01002853 JAC10745.1 CH916369 EDV93842.1 CH477801 EAT36085.1 KQ976574 KYM80153.1 ADTU01015323 CVRI01000006 CRK88002.1 KQ434777 KZC04191.1 GGFJ01007706 MBW56847.1 DQ440306 ABF18339.1 KQ414637 KOC66979.1 GANO01002731 JAB57140.1 GL439791 EFN66770.1 CCAG010018631 AY075510 AAL68318.1 GGFK01008331 MBW41652.1 GBXI01004881 JAD09411.1 ATLV01019092 KE525262 KFB43615.1 CH954181 EDV48777.1 AE014297 BT100172 AAF55370.1 ACY07077.1 AGB96032.1 CM000160 EDW96965.1 EDW96966.1 BT120319 AAF55369.2 ADC54210.1 AY070646 AAL48117.1 EDV48779.1 JXUM01032069 KQ560944 KXJ80243.1 CM000364 EDX12632.1 GAKP01006951 JAC52001.1 EDX12630.1 SPP83186.1 CH964272 EDW85169.1 GEZM01077729 JAV63193.1 KQ766607 OAD53614.1 GDKW01001411 JAI55184.1 CH480827 EDW44801.1 GAHY01000319 JAA77191.1 KQ971312 EEZ98669.1
BAM18412.1 AK405452 BAM20764.1 KQ460542 KPJ14122.1 GAIX01011519 JAA81041.1 GDQN01006373 JAT84681.1 AGBW02009968 OWR49461.1 KQ459579 KPI99487.1 NWSH01003045 PCG67051.1 JRES01000032 KNC34779.1 APCN01000816 AJWK01024414 AAAB01008987 EAA00991.4 GDAI01001568 JAI16035.1 GFDF01000039 JAV14045.1 AXCN02001485 CH940652 EDW58929.1 JXJN01004809 CH902617 EDV42311.1 EZ422781 ADD19057.1 UFQT01000415 SSX24106.1 AXCM01000086 UFQS01001255 UFQT01001255 SSX09952.1 SSX29675.1 GFDL01010679 JAV24366.1 AJVK01026700 ADMH02002101 ETN59171.1 KK107063 QOIP01000005 EZA60984.1 RLU23189.1 CH933806 EDW13686.1 KZ288253 PBC30935.1 JR047461 AEY60425.1 OUUW01000007 SPP83187.1 CM000070 EAL28216.1 CH479179 EDW25168.1 LJIG01022719 KRT79070.1 GAMC01001431 JAC05125.1 DS232197 EDS37189.1 GGFM01007409 MBW28160.1 GL888525 EGI59838.1 KQ980204 KYN17242.1 CP012526 ALC47888.1 KQ981387 KYN42024.1 KQ982294 KYQ58288.1 GAPW01002853 JAC10745.1 CH916369 EDV93842.1 CH477801 EAT36085.1 KQ976574 KYM80153.1 ADTU01015323 CVRI01000006 CRK88002.1 KQ434777 KZC04191.1 GGFJ01007706 MBW56847.1 DQ440306 ABF18339.1 KQ414637 KOC66979.1 GANO01002731 JAB57140.1 GL439791 EFN66770.1 CCAG010018631 AY075510 AAL68318.1 GGFK01008331 MBW41652.1 GBXI01004881 JAD09411.1 ATLV01019092 KE525262 KFB43615.1 CH954181 EDV48777.1 AE014297 BT100172 AAF55370.1 ACY07077.1 AGB96032.1 CM000160 EDW96965.1 EDW96966.1 BT120319 AAF55369.2 ADC54210.1 AY070646 AAL48117.1 EDV48779.1 JXUM01032069 KQ560944 KXJ80243.1 CM000364 EDX12632.1 GAKP01006951 JAC52001.1 EDX12630.1 SPP83186.1 CH964272 EDW85169.1 GEZM01077729 JAV63193.1 KQ766607 OAD53614.1 GDKW01001411 JAI55184.1 CH480827 EDW44801.1 GAHY01000319 JAA77191.1 KQ971312 EEZ98669.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000218220
UP000076407
+ More
UP000095300 UP000037069 UP000075903 UP000075902 UP000075840 UP000092461 UP000007062 UP000095301 UP000075882 UP000092443 UP000075881 UP000075886 UP000075884 UP000008792 UP000092460 UP000092445 UP000078200 UP000007801 UP000091820 UP000075883 UP000076408 UP000075920 UP000092462 UP000000673 UP000053097 UP000279307 UP000009192 UP000242457 UP000005203 UP000268350 UP000001819 UP000008744 UP000069272 UP000075885 UP000075900 UP000002320 UP000075901 UP000007755 UP000078492 UP000092553 UP000078541 UP000075809 UP000001070 UP000008820 UP000078540 UP000005205 UP000183832 UP000076502 UP000053825 UP000192221 UP000000311 UP000092444 UP000030765 UP000008711 UP000000803 UP000002282 UP000069940 UP000249989 UP000000304 UP000002358 UP000007798 UP000001292 UP000007266
UP000095300 UP000037069 UP000075903 UP000075902 UP000075840 UP000092461 UP000007062 UP000095301 UP000075882 UP000092443 UP000075881 UP000075886 UP000075884 UP000008792 UP000092460 UP000092445 UP000078200 UP000007801 UP000091820 UP000075883 UP000076408 UP000075920 UP000092462 UP000000673 UP000053097 UP000279307 UP000009192 UP000242457 UP000005203 UP000268350 UP000001819 UP000008744 UP000069272 UP000075885 UP000075900 UP000002320 UP000075901 UP000007755 UP000078492 UP000092553 UP000078541 UP000075809 UP000001070 UP000008820 UP000078540 UP000005205 UP000183832 UP000076502 UP000053825 UP000192221 UP000000311 UP000092444 UP000030765 UP000008711 UP000000803 UP000002282 UP000069940 UP000249989 UP000000304 UP000002358 UP000007798 UP000001292 UP000007266
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9JVM6
A0A2W1BGS9
A0A2H1WS25
I4DKH2
I4DS74
A0A0N0PCX0
+ More
S4NQJ4 A0A1E1WCU3 A0A212F6U5 A0A194Q1N1 A0A2A4J4Z3 A0A182XNZ1 A0A1I8Q7H9 A0A0L0CTR6 A0A182VEK3 A0A182TIY1 A0A182HK37 A0A1B0CR55 Q7PWZ6 A0A1I8MPL5 A0A2Y9D1G2 A0A0K8TQ66 A0A1L8E672 A0A1A9Y5Z0 A0A182JRG8 A0A182QE33 A0A182NR02 B4M5K3 A0A1B0AWJ2 A0A1A9ZFL9 A0A1A9UNW5 B3M2C0 D3TMX9 A0A1A9W598 A0A336M1A8 A0A182MGA1 A0A182YNB7 A0A182VW81 A0A336MI30 A0A1Q3F9Y8 A0A1B0D783 W5J5B0 A0A026X0R4 B4KBL9 A0A2A3EGP4 V9IH92 A0A088ATH4 A0A3B0JLP8 Q297I9 B4G4J7 A0A182FRZ4 A0A0T6AW84 A0A182PR69 A0A182RG10 W8CDS6 B0WYS9 A0A2M3ZI91 A0A182SAG7 F4X127 A0A195DWZ7 A0A0M4ERQ5 A0A195FQ25 A0A151XDE2 A0A023EP50 B4JHD5 Q16NY9 A0A195B6K9 A0A158NGY3 A0A1J1HKD6 A0A154NX87 A0A2M4BUW3 Q1HQY8 A0A0L7R806 A0A1W4VI77 U5ERN5 E2AII0 A0A1B0FFQ6 Q8SXY3 A0A2M4ALP4 A0A0A1XEQ9 A0A084W071 B3NZA5 Q9VEQ1 B4PM99 Q9VEQ2 Q8SZM8 B3NZA7 A0A182HAB0 B4QWE1 A0A034W8T7 B4QWD9 K7J067 A0A3B0KBX1 B4NKJ7 A0A1Y1KP61 A0A310S720 A0A0N7Z965 B4IBP3 R4FQM4 D6WAQ9
S4NQJ4 A0A1E1WCU3 A0A212F6U5 A0A194Q1N1 A0A2A4J4Z3 A0A182XNZ1 A0A1I8Q7H9 A0A0L0CTR6 A0A182VEK3 A0A182TIY1 A0A182HK37 A0A1B0CR55 Q7PWZ6 A0A1I8MPL5 A0A2Y9D1G2 A0A0K8TQ66 A0A1L8E672 A0A1A9Y5Z0 A0A182JRG8 A0A182QE33 A0A182NR02 B4M5K3 A0A1B0AWJ2 A0A1A9ZFL9 A0A1A9UNW5 B3M2C0 D3TMX9 A0A1A9W598 A0A336M1A8 A0A182MGA1 A0A182YNB7 A0A182VW81 A0A336MI30 A0A1Q3F9Y8 A0A1B0D783 W5J5B0 A0A026X0R4 B4KBL9 A0A2A3EGP4 V9IH92 A0A088ATH4 A0A3B0JLP8 Q297I9 B4G4J7 A0A182FRZ4 A0A0T6AW84 A0A182PR69 A0A182RG10 W8CDS6 B0WYS9 A0A2M3ZI91 A0A182SAG7 F4X127 A0A195DWZ7 A0A0M4ERQ5 A0A195FQ25 A0A151XDE2 A0A023EP50 B4JHD5 Q16NY9 A0A195B6K9 A0A158NGY3 A0A1J1HKD6 A0A154NX87 A0A2M4BUW3 Q1HQY8 A0A0L7R806 A0A1W4VI77 U5ERN5 E2AII0 A0A1B0FFQ6 Q8SXY3 A0A2M4ALP4 A0A0A1XEQ9 A0A084W071 B3NZA5 Q9VEQ1 B4PM99 Q9VEQ2 Q8SZM8 B3NZA7 A0A182HAB0 B4QWE1 A0A034W8T7 B4QWD9 K7J067 A0A3B0KBX1 B4NKJ7 A0A1Y1KP61 A0A310S720 A0A0N7Z965 B4IBP3 R4FQM4 D6WAQ9
Ontologies
GO
PANTHER
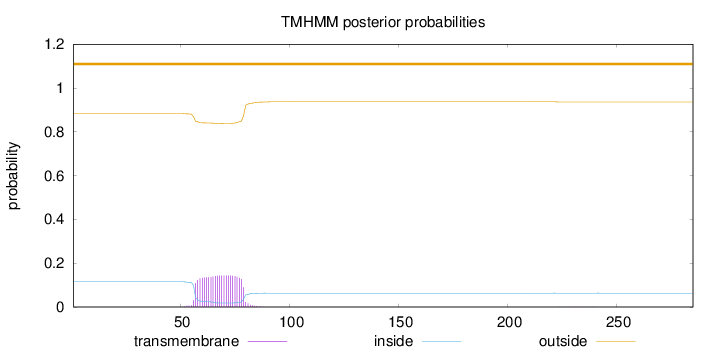
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.21621
Exp number, first 60 AAs:
0.54604
Total prob of N-in:
0.11636
outside
1 - 285
Population Genetic Test Statistics
Pi
54.842615
Theta
140.578492
Tajima's D
-1.025111
CLR
31.625283
CSRT
0.133843307834608
Interpretation
Possibly Positive selection
