Gene
KWMTBOMO03226
Pre Gene Modal
BGIBMGA013586
Annotation
PREDICTED:_synaptic_vesicle_2-related_protein-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.97
Sequence
CDS
ATGACCGTTGCTAAACATAACGAGGAGGAAACTATCGAAAGTGCCGTCGATAAAGCTGGTTTCGGTTTATACAGCATACTGTTAGTGATCTTGGTCGGCGTGTCGATTATATCGTTCGTATGCATCGGCTACAGCAGTACTATCCTGGTACCCACCAGCGCCTGCGAACTCCAGACCACCAGTAGCCAGCAAGGGATTCTAGCAGCTGGACCAGTTGTCGGTACCATAGTTGGCAGTTTAGTTTGGGGTTACCTAGCAGATACAAGAGGACGACGACTCATGCTGCTGGTATCATTGATGGGAAGCGCCATCATTAACATTGTGGCCAGCATATCTGTTAACTGGATTATGTTATTGATTTTGCAGTTCATTGCTGCTTTATTTGCGTCCGGTCTCTATTCGATGTCAATGTCGTTGTTGAGCGAGAGTGTCCCCATGGCGAAACGTAATTTGGTGATATTATTGGTGACCAGCATATTCTTGCTCTCACAAGGATTCATGGCGGCACTGGCCCTGCCAATAATTCCGTTGACTTTTTCATACTACATCTCTAGCTTGGACATCTATTGGAACTCTTGGAGGACCCTCGTGGTGGTCTACTCCATACCATGTATTTGTTCATTTATCTGCTTTTTATTCATGCAAGAGAGTCCTAAATTTGTCCACGCCATGGGAGATGAACAGAAGAGCTTAGAAATATTGAGGATAATTCATAAATATAATCATTTAGGTTCAAAAAAAGAGTTTCAGGTTTACCGAATAAAAAAGGCGGAGATGAACAAAAATGATACGCCGTCATCGTCAAAGGATCAAATAGTGCCGCTTTTTAGAGCGCCATTACTAAAGCCAACTATCATAATGACGTTATTGTACTTCTTTCAACAAGTTGGAGCGTTCATGGTGTGGCTTCCAACTATAGCCAACCAGTTCGTTCAAGTAACAGAAACAGGCGGGAACACAGATCTCACTCTTTGCGAAATCCTCAATAGTGAATTCGAAGTTTCAGTCAACCCTGACGTTGCTCCGTGCGCTCTCAACGAGACAGCACTGCTCATGGTTTTAGCCGTGGGTGCTTTACAGTGTGCATTCAATACAATTTTGAGTTTGATGGTAACTAAAATCGGAAGGCGCAATATTGTGATAGCCTTGGCGGGAATTTGTGGGCTGGCGGGGATTTTAGTGAACGTGGTACCAAATACCACGGGCAGCATTGTACTCTTCGGAATTTTCCTGATGGGGATAATCGTGATGGGTCTTTACACAGCTATCATCGTTGCGATTTTCCCTACACATTTACGGGCTATGGCCATCGCCTTACCACTGACTGCAAGCAGGATCACCACCTTCATCTCCATCCAGATCCTGAACTTGATGTTGGTGAACAACTGCGACGCTGGCTTCTATTTGTTTACATCAATATTTACATTATCGGCCGTTATAGCCGCCTTCCTACCAGACGATAGACGACTGCAGAAAGAAGTCAAAAATGAAACAGAAGCTAAGGCAGAAAATATAACTAAGTTGTAA
Protein
MTVAKHNEEETIESAVDKAGFGLYSILLVILVGVSIISFVCIGYSSTILVPTSACELQTTSSQQGILAAGPVVGTIVGSLVWGYLADTRGRRLMLLVSLMGSAIINIVASISVNWIMLLILQFIAALFASGLYSMSMSLLSESVPMAKRNLVILLVTSIFLLSQGFMAALALPIIPLTFSYYISSLDIYWNSWRTLVVVYSIPCICSFICFLFMQESPKFVHAMGDEQKSLEILRIIHKYNHLGSKKEFQVYRIKKAEMNKNDTPSSSKDQIVPLFRAPLLKPTIIMTLLYFFQQVGAFMVWLPTIANQFVQVTETGGNTDLTLCEILNSEFEVSVNPDVAPCALNETALLMVLAVGALQCAFNTILSLMVTKIGRRNIVIALAGICGLAGILVNVVPNTTGSIVLFGIFLMGIIVMGLYTAIIVAIFPTHLRAMAIALPLTASRITTFISIQILNLMLVNNCDAGFYLFTSIFTLSAVIAAFLPDDRRLQKEVKNETEAKAENITKL
Summary
Uniprot
A0A2A4JLT5
A0A2A4JMQ7
A0A2A4JLI7
A0A2H1WGI5
A0A0N1I9M8
A0A194Q3A9
+ More
A0A212F5P7 A0A2H1V0R1 A0A2H1VUK7 A0A2W1BZI9 A0A2A4JT62 H9JVL3 A0A194Q1R5 H9JVR6 A0A0N1I9N3 A0A0N1IPG1 A0A2H1WI98 A0A194Q2D2 A0A2H1V250 A0A2W1BWW5 A0A2H1W7K7 A0A2A4JX40 A0A2A4JLN1 A0A212EZN8 A0A2A4JKW1 A0A2H1WYY1 A0A212FI93 A0A212EY43 A0A194PP30 A0A194QN61 A0A2W1BUU8 A0A194Q1T6 A0A194Q1T1 A0A2H1W801 A0A1Q3FLR3 A0A194PJI3 A0A212EQC0 H9JVM2 A0A182QCG3 A0A212EY03 A0A212EYD8 B0W415 A0A1Q3FNK2 A0A1B0CB10 A0A182NE16 U5ETH9 F5HLT0 A0A1L8EFY3 H9JVL1 A0A1L8EFQ6 A0A0N0PCN2 A0A1L8EFU2 A0A1I8P1U8 A0A1I8P1R5 Q17HA7 B0WQD3 A0A084W7Z4 D6WB98 A0A1J1HSM7 A0A1Q3FH96 A0A1B6I2U3 A0A1L8DUU9 A0A2H1X0I9 A0A1L8DEL2 A0A182GWL7 A0A182W1I9 A0A182MH17 A0A182H160 A0A2A4K5Q7 A0A067RD65 A0A2M4CS78 A0A182YGM4 A0A182N5S5 A0A1S4EXV1 Q17M95 A0A1Q3FM89 A0A0L7KD47 A0A212F5C2 A0A2M4CS45 B0WKE4 A0A2M4BKA1 T1PCW5 A0A1I8MVY3 A0A1L8DEG9 A0A1I8MVX6 A0A182GKA8 A0A182FPU8 A0A182P1Q5 A0A2M4BKB0 Q7QB66 A0A1S4G380 A0A2M4BKT9 A0A194Q1Q2 A0A182RYM7
A0A212F5P7 A0A2H1V0R1 A0A2H1VUK7 A0A2W1BZI9 A0A2A4JT62 H9JVL3 A0A194Q1R5 H9JVR6 A0A0N1I9N3 A0A0N1IPG1 A0A2H1WI98 A0A194Q2D2 A0A2H1V250 A0A2W1BWW5 A0A2H1W7K7 A0A2A4JX40 A0A2A4JLN1 A0A212EZN8 A0A2A4JKW1 A0A2H1WYY1 A0A212FI93 A0A212EY43 A0A194PP30 A0A194QN61 A0A2W1BUU8 A0A194Q1T6 A0A194Q1T1 A0A2H1W801 A0A1Q3FLR3 A0A194PJI3 A0A212EQC0 H9JVM2 A0A182QCG3 A0A212EY03 A0A212EYD8 B0W415 A0A1Q3FNK2 A0A1B0CB10 A0A182NE16 U5ETH9 F5HLT0 A0A1L8EFY3 H9JVL1 A0A1L8EFQ6 A0A0N0PCN2 A0A1L8EFU2 A0A1I8P1U8 A0A1I8P1R5 Q17HA7 B0WQD3 A0A084W7Z4 D6WB98 A0A1J1HSM7 A0A1Q3FH96 A0A1B6I2U3 A0A1L8DUU9 A0A2H1X0I9 A0A1L8DEL2 A0A182GWL7 A0A182W1I9 A0A182MH17 A0A182H160 A0A2A4K5Q7 A0A067RD65 A0A2M4CS78 A0A182YGM4 A0A182N5S5 A0A1S4EXV1 Q17M95 A0A1Q3FM89 A0A0L7KD47 A0A212F5C2 A0A2M4CS45 B0WKE4 A0A2M4BKA1 T1PCW5 A0A1I8MVY3 A0A1L8DEG9 A0A1I8MVX6 A0A182GKA8 A0A182FPU8 A0A182P1Q5 A0A2M4BKB0 Q7QB66 A0A1S4G380 A0A2M4BKT9 A0A194Q1Q2 A0A182RYM7
Pubmed
EMBL
NWSH01001089
PCG72678.1
PCG72680.1
PCG72679.1
ODYU01008516
SOQ52183.1
+ More
KQ460542 KPJ14130.1 KQ459579 KPI99494.1 AGBW02010155 OWR49057.1 ODYU01000151 SOQ34435.1 ODYU01004537 SOQ44529.1 KZ149911 PZC78136.1 NWSH01000659 PCG74966.1 BABH01032453 KPI99491.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 KPJ14146.1 KPJ14127.1 SOQ52184.1 KPI99493.1 SOQ34432.1 PZC78135.1 ODYU01006861 SOQ49085.1 NWSH01000457 PCG76268.1 PCG72686.1 AGBW02011241 OWR46942.1 PCG72687.1 ODYU01012071 SOQ58176.1 AGBW02008417 OWR53448.1 AGBW02011638 OWR46364.1 KQ459603 KPI92885.1 KQ461194 KPJ06947.1 PZC78141.1 KPI99511.1 KPI99506.1 SOQ49086.1 GFDL01006485 JAV28560.1 KPI92884.1 AGBW02013272 OWR43679.1 BABH01032396 BABH01032397 BABH01032398 BABH01032399 AXCN02002212 AGBW02011653 OWR46341.1 AGBW02011531 OWR46513.1 DS231834 EDS32608.1 GFDL01005870 JAV29175.1 AJWK01004723 GANO01004365 JAB55506.1 AAAB01008952 EGK97241.1 GFDG01001245 JAV17554.1 BABH01032458 GFDG01001242 JAV17557.1 KPJ14131.1 GFDG01001241 JAV17558.1 CH477251 EAT46035.1 DS232039 EDS32804.1 ATLV01021326 KE525316 KFB46338.1 KQ971312 EEZ97917.1 CVRI01000020 CRK91005.1 GFDL01008118 JAV26927.1 GECU01026455 JAS81251.1 GFDF01003888 JAV10196.1 ODYU01012422 SOQ58716.1 GFDF01009181 JAV04903.1 JXUM01093536 KQ564063 KXJ72850.1 AXCM01000270 JXUM01102795 JXUM01102796 KQ564753 KXJ71786.1 NWSH01000098 PCG79585.1 KK852543 KDR21687.1 GGFL01003985 MBW68163.1 CH477207 EAT47854.1 GFDL01006432 JAV28613.1 JTDY01010086 KOB61253.1 AGBW02010212 OWR48924.1 GGFL01003986 MBW68164.1 DS231971 EDS29763.1 GGFJ01004344 MBW53485.1 KA646499 AFP61128.1 GFDF01009225 JAV04859.1 JXUM01069579 KQ562558 KXJ75630.1 GGFJ01004346 MBW53487.1 AAAB01008880 EAA08657.4 EGK97004.1 GGFJ01004430 MBW53571.1 KPI99507.1
KQ460542 KPJ14130.1 KQ459579 KPI99494.1 AGBW02010155 OWR49057.1 ODYU01000151 SOQ34435.1 ODYU01004537 SOQ44529.1 KZ149911 PZC78136.1 NWSH01000659 PCG74966.1 BABH01032453 KPI99491.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 KPJ14146.1 KPJ14127.1 SOQ52184.1 KPI99493.1 SOQ34432.1 PZC78135.1 ODYU01006861 SOQ49085.1 NWSH01000457 PCG76268.1 PCG72686.1 AGBW02011241 OWR46942.1 PCG72687.1 ODYU01012071 SOQ58176.1 AGBW02008417 OWR53448.1 AGBW02011638 OWR46364.1 KQ459603 KPI92885.1 KQ461194 KPJ06947.1 PZC78141.1 KPI99511.1 KPI99506.1 SOQ49086.1 GFDL01006485 JAV28560.1 KPI92884.1 AGBW02013272 OWR43679.1 BABH01032396 BABH01032397 BABH01032398 BABH01032399 AXCN02002212 AGBW02011653 OWR46341.1 AGBW02011531 OWR46513.1 DS231834 EDS32608.1 GFDL01005870 JAV29175.1 AJWK01004723 GANO01004365 JAB55506.1 AAAB01008952 EGK97241.1 GFDG01001245 JAV17554.1 BABH01032458 GFDG01001242 JAV17557.1 KPJ14131.1 GFDG01001241 JAV17558.1 CH477251 EAT46035.1 DS232039 EDS32804.1 ATLV01021326 KE525316 KFB46338.1 KQ971312 EEZ97917.1 CVRI01000020 CRK91005.1 GFDL01008118 JAV26927.1 GECU01026455 JAS81251.1 GFDF01003888 JAV10196.1 ODYU01012422 SOQ58716.1 GFDF01009181 JAV04903.1 JXUM01093536 KQ564063 KXJ72850.1 AXCM01000270 JXUM01102795 JXUM01102796 KQ564753 KXJ71786.1 NWSH01000098 PCG79585.1 KK852543 KDR21687.1 GGFL01003985 MBW68163.1 CH477207 EAT47854.1 GFDL01006432 JAV28613.1 JTDY01010086 KOB61253.1 AGBW02010212 OWR48924.1 GGFL01003986 MBW68164.1 DS231971 EDS29763.1 GGFJ01004344 MBW53485.1 KA646499 AFP61128.1 GFDF01009225 JAV04859.1 JXUM01069579 KQ562558 KXJ75630.1 GGFJ01004346 MBW53487.1 AAAB01008880 EAA08657.4 EGK97004.1 GGFJ01004430 MBW53571.1 KPI99507.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000075886
+ More
UP000002320 UP000092461 UP000075884 UP000007062 UP000095300 UP000008820 UP000030765 UP000007266 UP000183832 UP000069940 UP000249989 UP000075920 UP000075883 UP000027135 UP000076408 UP000037510 UP000095301 UP000069272 UP000075885 UP000075900
UP000002320 UP000092461 UP000075884 UP000007062 UP000095300 UP000008820 UP000030765 UP000007266 UP000183832 UP000069940 UP000249989 UP000075920 UP000075883 UP000027135 UP000076408 UP000037510 UP000095301 UP000069272 UP000075885 UP000075900
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JLT5
A0A2A4JMQ7
A0A2A4JLI7
A0A2H1WGI5
A0A0N1I9M8
A0A194Q3A9
+ More
A0A212F5P7 A0A2H1V0R1 A0A2H1VUK7 A0A2W1BZI9 A0A2A4JT62 H9JVL3 A0A194Q1R5 H9JVR6 A0A0N1I9N3 A0A0N1IPG1 A0A2H1WI98 A0A194Q2D2 A0A2H1V250 A0A2W1BWW5 A0A2H1W7K7 A0A2A4JX40 A0A2A4JLN1 A0A212EZN8 A0A2A4JKW1 A0A2H1WYY1 A0A212FI93 A0A212EY43 A0A194PP30 A0A194QN61 A0A2W1BUU8 A0A194Q1T6 A0A194Q1T1 A0A2H1W801 A0A1Q3FLR3 A0A194PJI3 A0A212EQC0 H9JVM2 A0A182QCG3 A0A212EY03 A0A212EYD8 B0W415 A0A1Q3FNK2 A0A1B0CB10 A0A182NE16 U5ETH9 F5HLT0 A0A1L8EFY3 H9JVL1 A0A1L8EFQ6 A0A0N0PCN2 A0A1L8EFU2 A0A1I8P1U8 A0A1I8P1R5 Q17HA7 B0WQD3 A0A084W7Z4 D6WB98 A0A1J1HSM7 A0A1Q3FH96 A0A1B6I2U3 A0A1L8DUU9 A0A2H1X0I9 A0A1L8DEL2 A0A182GWL7 A0A182W1I9 A0A182MH17 A0A182H160 A0A2A4K5Q7 A0A067RD65 A0A2M4CS78 A0A182YGM4 A0A182N5S5 A0A1S4EXV1 Q17M95 A0A1Q3FM89 A0A0L7KD47 A0A212F5C2 A0A2M4CS45 B0WKE4 A0A2M4BKA1 T1PCW5 A0A1I8MVY3 A0A1L8DEG9 A0A1I8MVX6 A0A182GKA8 A0A182FPU8 A0A182P1Q5 A0A2M4BKB0 Q7QB66 A0A1S4G380 A0A2M4BKT9 A0A194Q1Q2 A0A182RYM7
A0A212F5P7 A0A2H1V0R1 A0A2H1VUK7 A0A2W1BZI9 A0A2A4JT62 H9JVL3 A0A194Q1R5 H9JVR6 A0A0N1I9N3 A0A0N1IPG1 A0A2H1WI98 A0A194Q2D2 A0A2H1V250 A0A2W1BWW5 A0A2H1W7K7 A0A2A4JX40 A0A2A4JLN1 A0A212EZN8 A0A2A4JKW1 A0A2H1WYY1 A0A212FI93 A0A212EY43 A0A194PP30 A0A194QN61 A0A2W1BUU8 A0A194Q1T6 A0A194Q1T1 A0A2H1W801 A0A1Q3FLR3 A0A194PJI3 A0A212EQC0 H9JVM2 A0A182QCG3 A0A212EY03 A0A212EYD8 B0W415 A0A1Q3FNK2 A0A1B0CB10 A0A182NE16 U5ETH9 F5HLT0 A0A1L8EFY3 H9JVL1 A0A1L8EFQ6 A0A0N0PCN2 A0A1L8EFU2 A0A1I8P1U8 A0A1I8P1R5 Q17HA7 B0WQD3 A0A084W7Z4 D6WB98 A0A1J1HSM7 A0A1Q3FH96 A0A1B6I2U3 A0A1L8DUU9 A0A2H1X0I9 A0A1L8DEL2 A0A182GWL7 A0A182W1I9 A0A182MH17 A0A182H160 A0A2A4K5Q7 A0A067RD65 A0A2M4CS78 A0A182YGM4 A0A182N5S5 A0A1S4EXV1 Q17M95 A0A1Q3FM89 A0A0L7KD47 A0A212F5C2 A0A2M4CS45 B0WKE4 A0A2M4BKA1 T1PCW5 A0A1I8MVY3 A0A1L8DEG9 A0A1I8MVX6 A0A182GKA8 A0A182FPU8 A0A182P1Q5 A0A2M4BKB0 Q7QB66 A0A1S4G380 A0A2M4BKT9 A0A194Q1Q2 A0A182RYM7
Ontologies
GO
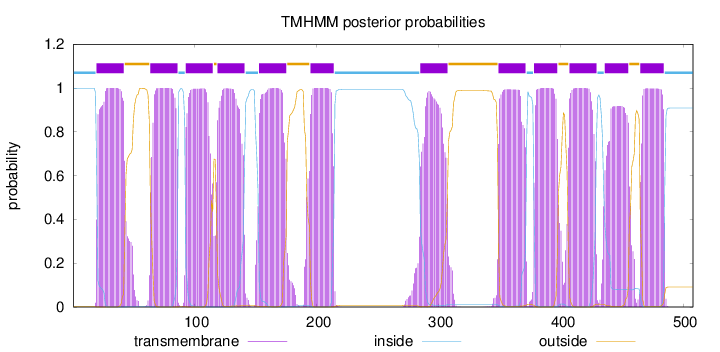
Topology
Length:
508
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
262.57127
Exp number, first 60 AAs:
24.6515
Total prob of N-in:
0.99882
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 63
TMhelix
64 - 86
inside
87 - 92
TMhelix
93 - 115
outside
116 - 118
TMhelix
119 - 141
inside
142 - 152
TMhelix
153 - 175
outside
176 - 194
TMhelix
195 - 214
inside
215 - 284
TMhelix
285 - 307
outside
308 - 348
TMhelix
349 - 371
inside
372 - 377
TMhelix
378 - 397
outside
398 - 406
TMhelix
407 - 429
inside
430 - 435
TMhelix
436 - 455
outside
456 - 464
TMhelix
465 - 484
inside
485 - 508
Population Genetic Test Statistics
Pi
162.527728
Theta
161.159252
Tajima's D
0.021734
CLR
0.267045
CSRT
0.371181440927954
Interpretation
Uncertain
