Gene
KWMTBOMO03224
Pre Gene Modal
BGIBMGA013585
Annotation
putative_SV2-like_protein_1?_partial_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 2.692
Sequence
CDS
ATGGAATTTTCATATCCCATTAATTTCCTAAACATCACATTCCGTTCCTGGCGTCTTTTGATCCTCGTCCTGGCAGTGCCATGTGCGATCACAGCTGGCCTGCTGCAGTTCTTCCACGAGAGCCCCAAGTTCCTGACCTCTCAGGGTCGATACGATGAAGCTCTGGTTGTGCTGAAGAAGATTTACGCCTGTAATACTGGAGATAGAGCCGAAAATTTTCCGGTCAAAAAACTCATTCCGGACAACGAGGACCAGACTGCCGAAGCCCAGGATTCGTTTCTCAAGTCCCTGTGGCATCAAACCGTGTCGCTGTTCAAGCCTCCATTGCTCAAGGATACGATGAGACTATTCTATTTGGTCATTATCATTTATATGACGTAA
Protein
MEFSYPINFLNITFRSWRLLILVLAVPCAITAGLLQFFHESPKFLTSQGRYDEALVVLKKIYACNTGDRAENFPVKKLIPDNEDQTAEAQDSFLKSLWHQTVSLFKPPLLKDTMRLFYLVIIIYMT
Summary
Uniprot
H9JVM2
A0A2A4JL38
A0A0L7L435
A0A0N1IG89
A0A194Q2D2
A0A212EY03
+ More
A0A2H1WI98 A0A2H1W801 A0A0L7KRD9 A0A2H1V0R1 A0A2H1V6C3 A0A194Q800 A0A2W1BUU8 A0A2W1BGE8 A0A0L7KD47 A0A2H1VEB4 A0A0N1PIZ7 A0A0N1IPG1 A0A2W1BZI9 A0A2H1VUK7 A0A2H1V250 A0A194Q1R5 A0A194Q1T6 H9JVR7 A0A1J1HSM7 A0A2A4JT62 A0A212EQC0 A0A0N0PCK2 A0A212EY43 A0A2H1W7K7 A0A0N0PCN2 A0A2H1WJU4 A0A2A4IYX6 A0A2A4JLN1 A0A2A4JKW1 H9JVL3 A0A2W1BQE4 A0A194Q1Q2 A0A2A4JAH3 H9JVR5 A0A0L7LS59 A0A0N1I9N3 H9JV72 A0A194Q1T1 A0A0L7LHZ8 H9JVS5 H9JVR6 H9JVL1 A0A212FI93 A0A154NYA8 A0A0L7KVC0 A0A212EZN8 A0A2H1WU82 A0A2H1VZK7 T1GFD6 Q17HA7 A0A2A4JX40 A0A212F5C2 A0A0L7QLM7 E2ATB0 A0A170Y9B6 A0A2W1BVQ5 A0A310SDB3 A0A2A3EPR0 A0A194Q7W0 A0A195E6G9 A0A212EYD8 A0A2H1WYY1 S4Q006 A0A182GWL7 A0A195CY83 A0A195F2P9 A0A151XCA7 A0A2H1X0I9 A0A195B1S0 A0A158NLE1 E2BMY6 A0A2H1WYT6 B0WDR9 N6T4U4 E2BMY5 A0A1B6MD06 A0A2J7RAM5 A0A0A9XV82 V5GNJ1 Q178J2 A0A2A4J999 A0A2J7RAM3 F4WW57 R4WKX5 A0A1Q3FMH0 A0A084WQ83 A0A023EU38 A0A1B6DTT2 A0A1B6JMS4 A0A146KQ36 A0A0A9YRF3 A0A182VKS7 T1GER7 W5J8T8
A0A2H1WI98 A0A2H1W801 A0A0L7KRD9 A0A2H1V0R1 A0A2H1V6C3 A0A194Q800 A0A2W1BUU8 A0A2W1BGE8 A0A0L7KD47 A0A2H1VEB4 A0A0N1PIZ7 A0A0N1IPG1 A0A2W1BZI9 A0A2H1VUK7 A0A2H1V250 A0A194Q1R5 A0A194Q1T6 H9JVR7 A0A1J1HSM7 A0A2A4JT62 A0A212EQC0 A0A0N0PCK2 A0A212EY43 A0A2H1W7K7 A0A0N0PCN2 A0A2H1WJU4 A0A2A4IYX6 A0A2A4JLN1 A0A2A4JKW1 H9JVL3 A0A2W1BQE4 A0A194Q1Q2 A0A2A4JAH3 H9JVR5 A0A0L7LS59 A0A0N1I9N3 H9JV72 A0A194Q1T1 A0A0L7LHZ8 H9JVS5 H9JVR6 H9JVL1 A0A212FI93 A0A154NYA8 A0A0L7KVC0 A0A212EZN8 A0A2H1WU82 A0A2H1VZK7 T1GFD6 Q17HA7 A0A2A4JX40 A0A212F5C2 A0A0L7QLM7 E2ATB0 A0A170Y9B6 A0A2W1BVQ5 A0A310SDB3 A0A2A3EPR0 A0A194Q7W0 A0A195E6G9 A0A212EYD8 A0A2H1WYY1 S4Q006 A0A182GWL7 A0A195CY83 A0A195F2P9 A0A151XCA7 A0A2H1X0I9 A0A195B1S0 A0A158NLE1 E2BMY6 A0A2H1WYT6 B0WDR9 N6T4U4 E2BMY5 A0A1B6MD06 A0A2J7RAM5 A0A0A9XV82 V5GNJ1 Q178J2 A0A2A4J999 A0A2J7RAM3 F4WW57 R4WKX5 A0A1Q3FMH0 A0A084WQ83 A0A023EU38 A0A1B6DTT2 A0A1B6JMS4 A0A146KQ36 A0A0A9YRF3 A0A182VKS7 T1GER7 W5J8T8
Pubmed
EMBL
BABH01032396
BABH01032397
BABH01032398
BABH01032399
NWSH01001089
PCG72681.1
+ More
JTDY01003063 KOB70227.1 KQ460542 KPJ14129.1 KQ459579 KPI99493.1 AGBW02011653 OWR46341.1 ODYU01008516 SOQ52184.1 ODYU01006861 SOQ49086.1 JTDY01006887 KOB65616.1 ODYU01000151 SOQ34435.1 ODYU01000910 SOQ36383.1 KPI99535.1 KZ149911 PZC78141.1 KZ150202 PZC72277.1 JTDY01010086 KOB61253.1 ODYU01001846 SOQ38614.1 KPJ14149.1 KPJ14127.1 PZC78136.1 ODYU01004537 SOQ44529.1 SOQ34432.1 KPI99491.1 KPI99511.1 BABH01032426 BABH01032427 BABH01032428 BABH01032429 CVRI01000020 CRK91005.1 NWSH01000659 PCG74966.1 AGBW02013272 OWR43679.1 KPJ14144.1 AGBW02011638 OWR46364.1 SOQ49085.1 KPJ14131.1 ODYU01009141 SOQ53353.1 NWSH01005206 PCG64352.1 PCG72686.1 PCG72687.1 BABH01032453 KZ149979 PZC75844.1 KPI99507.1 NWSH01002380 PCG68430.1 BABH01032417 JTDY01000209 KOB78282.1 KPJ14146.1 BABH01037389 KPI99506.1 JTDY01001109 KOB74851.1 BABH01032451 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 BABH01032458 AGBW02008417 OWR53448.1 KQ434773 KZC04008.1 JTDY01005416 KOB67031.1 AGBW02011241 OWR46942.1 ODYU01011059 SOQ56557.1 ODYU01005122 SOQ45674.1 CAQQ02025659 CAQQ02025660 CAQQ02025661 CH477251 EAT46035.1 NWSH01000457 PCG76268.1 AGBW02010212 OWR48924.1 KQ414914 KOC59533.1 GL442545 EFN63355.1 GEMB01003525 JAR99692.1 PZC75843.1 KQ775195 OAD52273.1 KZ288197 PBC33718.1 KPI99495.1 KQ979568 KYN20768.1 AGBW02011531 OWR46513.1 ODYU01012071 SOQ58176.1 GAIX01000623 JAA91937.1 JXUM01093536 KQ564063 KXJ72850.1 KQ977115 KYN05613.1 KQ981864 KYN34369.1 KQ982314 KYQ58026.1 ODYU01012422 SOQ58716.1 KQ976662 KYM78421.1 ADTU01019324 GL449382 EFN82890.1 ODYU01012108 SOQ58229.1 DS231900 EDS44822.1 APGK01052662 KB741213 ENN72643.1 EFN82889.1 GEBQ01006218 JAT33759.1 NEVH01006567 PNF37880.1 GBHO01023261 GBHO01020300 GBHO01020298 GBHO01003596 JAG20343.1 JAG23304.1 JAG23306.1 JAG40008.1 GALX01002772 JAB65694.1 CH477362 EAT42650.1 PCG68429.1 PNF37878.1 GL888404 EGI61528.1 AK418278 BAN21490.1 GFDL01006323 JAV28722.1 ATLV01025286 KE525379 KFB52377.1 GAPW01000710 JAC12888.1 GEDC01008249 JAS29049.1 GECU01007523 JAT00184.1 GDHC01020470 JAP98158.1 GBHO01034941 GBHO01034937 GBHO01034935 GBHO01009383 JAG08663.1 JAG08667.1 JAG08669.1 JAG34221.1 CAQQ02083380 CAQQ02083381 ADMH02001852 ETN60872.1
JTDY01003063 KOB70227.1 KQ460542 KPJ14129.1 KQ459579 KPI99493.1 AGBW02011653 OWR46341.1 ODYU01008516 SOQ52184.1 ODYU01006861 SOQ49086.1 JTDY01006887 KOB65616.1 ODYU01000151 SOQ34435.1 ODYU01000910 SOQ36383.1 KPI99535.1 KZ149911 PZC78141.1 KZ150202 PZC72277.1 JTDY01010086 KOB61253.1 ODYU01001846 SOQ38614.1 KPJ14149.1 KPJ14127.1 PZC78136.1 ODYU01004537 SOQ44529.1 SOQ34432.1 KPI99491.1 KPI99511.1 BABH01032426 BABH01032427 BABH01032428 BABH01032429 CVRI01000020 CRK91005.1 NWSH01000659 PCG74966.1 AGBW02013272 OWR43679.1 KPJ14144.1 AGBW02011638 OWR46364.1 SOQ49085.1 KPJ14131.1 ODYU01009141 SOQ53353.1 NWSH01005206 PCG64352.1 PCG72686.1 PCG72687.1 BABH01032453 KZ149979 PZC75844.1 KPI99507.1 NWSH01002380 PCG68430.1 BABH01032417 JTDY01000209 KOB78282.1 KPJ14146.1 BABH01037389 KPI99506.1 JTDY01001109 KOB74851.1 BABH01032451 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 BABH01032458 AGBW02008417 OWR53448.1 KQ434773 KZC04008.1 JTDY01005416 KOB67031.1 AGBW02011241 OWR46942.1 ODYU01011059 SOQ56557.1 ODYU01005122 SOQ45674.1 CAQQ02025659 CAQQ02025660 CAQQ02025661 CH477251 EAT46035.1 NWSH01000457 PCG76268.1 AGBW02010212 OWR48924.1 KQ414914 KOC59533.1 GL442545 EFN63355.1 GEMB01003525 JAR99692.1 PZC75843.1 KQ775195 OAD52273.1 KZ288197 PBC33718.1 KPI99495.1 KQ979568 KYN20768.1 AGBW02011531 OWR46513.1 ODYU01012071 SOQ58176.1 GAIX01000623 JAA91937.1 JXUM01093536 KQ564063 KXJ72850.1 KQ977115 KYN05613.1 KQ981864 KYN34369.1 KQ982314 KYQ58026.1 ODYU01012422 SOQ58716.1 KQ976662 KYM78421.1 ADTU01019324 GL449382 EFN82890.1 ODYU01012108 SOQ58229.1 DS231900 EDS44822.1 APGK01052662 KB741213 ENN72643.1 EFN82889.1 GEBQ01006218 JAT33759.1 NEVH01006567 PNF37880.1 GBHO01023261 GBHO01020300 GBHO01020298 GBHO01003596 JAG20343.1 JAG23304.1 JAG23306.1 JAG40008.1 GALX01002772 JAB65694.1 CH477362 EAT42650.1 PCG68429.1 PNF37878.1 GL888404 EGI61528.1 AK418278 BAN21490.1 GFDL01006323 JAV28722.1 ATLV01025286 KE525379 KFB52377.1 GAPW01000710 JAC12888.1 GEDC01008249 JAS29049.1 GECU01007523 JAT00184.1 GDHC01020470 JAP98158.1 GBHO01034941 GBHO01034937 GBHO01034935 GBHO01009383 JAG08663.1 JAG08667.1 JAG08669.1 JAG34221.1 CAQQ02083380 CAQQ02083381 ADMH02001852 ETN60872.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000183832 UP000076502 UP000015102 UP000008820 UP000053825 UP000000311 UP000242457 UP000078492 UP000069940 UP000249989 UP000078542 UP000078541 UP000075809 UP000078540 UP000005205 UP000008237 UP000002320 UP000019118 UP000235965 UP000007755 UP000030765 UP000075903 UP000000673
UP000183832 UP000076502 UP000015102 UP000008820 UP000053825 UP000000311 UP000242457 UP000078492 UP000069940 UP000249989 UP000078542 UP000078541 UP000075809 UP000078540 UP000005205 UP000008237 UP000002320 UP000019118 UP000235965 UP000007755 UP000030765 UP000075903 UP000000673
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JVM2
A0A2A4JL38
A0A0L7L435
A0A0N1IG89
A0A194Q2D2
A0A212EY03
+ More
A0A2H1WI98 A0A2H1W801 A0A0L7KRD9 A0A2H1V0R1 A0A2H1V6C3 A0A194Q800 A0A2W1BUU8 A0A2W1BGE8 A0A0L7KD47 A0A2H1VEB4 A0A0N1PIZ7 A0A0N1IPG1 A0A2W1BZI9 A0A2H1VUK7 A0A2H1V250 A0A194Q1R5 A0A194Q1T6 H9JVR7 A0A1J1HSM7 A0A2A4JT62 A0A212EQC0 A0A0N0PCK2 A0A212EY43 A0A2H1W7K7 A0A0N0PCN2 A0A2H1WJU4 A0A2A4IYX6 A0A2A4JLN1 A0A2A4JKW1 H9JVL3 A0A2W1BQE4 A0A194Q1Q2 A0A2A4JAH3 H9JVR5 A0A0L7LS59 A0A0N1I9N3 H9JV72 A0A194Q1T1 A0A0L7LHZ8 H9JVS5 H9JVR6 H9JVL1 A0A212FI93 A0A154NYA8 A0A0L7KVC0 A0A212EZN8 A0A2H1WU82 A0A2H1VZK7 T1GFD6 Q17HA7 A0A2A4JX40 A0A212F5C2 A0A0L7QLM7 E2ATB0 A0A170Y9B6 A0A2W1BVQ5 A0A310SDB3 A0A2A3EPR0 A0A194Q7W0 A0A195E6G9 A0A212EYD8 A0A2H1WYY1 S4Q006 A0A182GWL7 A0A195CY83 A0A195F2P9 A0A151XCA7 A0A2H1X0I9 A0A195B1S0 A0A158NLE1 E2BMY6 A0A2H1WYT6 B0WDR9 N6T4U4 E2BMY5 A0A1B6MD06 A0A2J7RAM5 A0A0A9XV82 V5GNJ1 Q178J2 A0A2A4J999 A0A2J7RAM3 F4WW57 R4WKX5 A0A1Q3FMH0 A0A084WQ83 A0A023EU38 A0A1B6DTT2 A0A1B6JMS4 A0A146KQ36 A0A0A9YRF3 A0A182VKS7 T1GER7 W5J8T8
A0A2H1WI98 A0A2H1W801 A0A0L7KRD9 A0A2H1V0R1 A0A2H1V6C3 A0A194Q800 A0A2W1BUU8 A0A2W1BGE8 A0A0L7KD47 A0A2H1VEB4 A0A0N1PIZ7 A0A0N1IPG1 A0A2W1BZI9 A0A2H1VUK7 A0A2H1V250 A0A194Q1R5 A0A194Q1T6 H9JVR7 A0A1J1HSM7 A0A2A4JT62 A0A212EQC0 A0A0N0PCK2 A0A212EY43 A0A2H1W7K7 A0A0N0PCN2 A0A2H1WJU4 A0A2A4IYX6 A0A2A4JLN1 A0A2A4JKW1 H9JVL3 A0A2W1BQE4 A0A194Q1Q2 A0A2A4JAH3 H9JVR5 A0A0L7LS59 A0A0N1I9N3 H9JV72 A0A194Q1T1 A0A0L7LHZ8 H9JVS5 H9JVR6 H9JVL1 A0A212FI93 A0A154NYA8 A0A0L7KVC0 A0A212EZN8 A0A2H1WU82 A0A2H1VZK7 T1GFD6 Q17HA7 A0A2A4JX40 A0A212F5C2 A0A0L7QLM7 E2ATB0 A0A170Y9B6 A0A2W1BVQ5 A0A310SDB3 A0A2A3EPR0 A0A194Q7W0 A0A195E6G9 A0A212EYD8 A0A2H1WYY1 S4Q006 A0A182GWL7 A0A195CY83 A0A195F2P9 A0A151XCA7 A0A2H1X0I9 A0A195B1S0 A0A158NLE1 E2BMY6 A0A2H1WYT6 B0WDR9 N6T4U4 E2BMY5 A0A1B6MD06 A0A2J7RAM5 A0A0A9XV82 V5GNJ1 Q178J2 A0A2A4J999 A0A2J7RAM3 F4WW57 R4WKX5 A0A1Q3FMH0 A0A084WQ83 A0A023EU38 A0A1B6DTT2 A0A1B6JMS4 A0A146KQ36 A0A0A9YRF3 A0A182VKS7 T1GER7 W5J8T8
Ontologies
GO
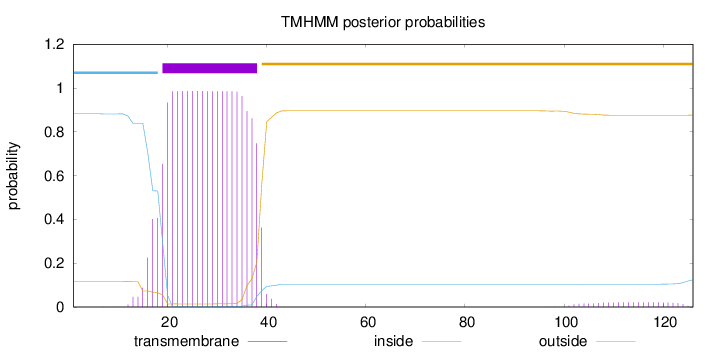
Topology
Length:
126
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.99016
Exp number, first 60 AAs:
20.5444
Total prob of N-in:
0.88256
POSSIBLE N-term signal
sequence
inside
1 - 18
TMhelix
19 - 38
outside
39 - 126
Population Genetic Test Statistics
Pi
46.721276
Theta
123.01255
Tajima's D
-1.011814
CLR
0.680283
CSRT
0.137793110344483
Interpretation
Uncertain
