Gene
KWMTBOMO03223
Annotation
PREDICTED:_synaptic_vesicle_glycoprotein_2C-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.44
Sequence
CDS
ATGAAGGAGAAAAATATGGCTTTAAAAGATGTGAAGGAAAAATCTGATTTACCACAGAAAGTACTGGAAAATGTGGATCTAGAAGACGCCTTAGATCTGGCAGGCGCGGGCCGGTATCAATTAGTTCACACTGCACTGATGCTGTTCTCAACGACGGCTGCGATATTAGAAATGATCGGAGTGGCTTTCGTGCTCCCAGCCGCGACATGCGACCTTCAATTATCTGATAAACTCAGAGGAATCATCACTTCGATACCTAACATAGGCGTCATCCTCACGGCGGCGCTGTGGGGTAGAGCATCGGACACTTTGGGCCGGAAGCCAGCAATGATTGCGTCTGCTTCGATGGGCGGCTTCGTTGGACTCCTGGCTGCGTTCATGCCTAATTTGTTAGCGTTCAGCATCTGCAAGTTTTGTGGTTCACTTTTTCTGTCAAGCGCGTCATCTTTGGGTTTCGCGTATATTTGCGAGATGCTGCCAGTGAAGAGTCGTGACAGAACCGTTCTGATCTGCAATGGATTAGTGATGATACTCTCCACACTATGTCCTGGTAAGCTTCCACCAGCATTTTTTTTATTGCCCTTATAG
Protein
MKEKNMALKDVKEKSDLPQKVLENVDLEDALDLAGAGRYQLVHTALMLFSTTAAILEMIGVAFVLPAATCDLQLSDKLRGIITSIPNIGVILTAALWGRASDTLGRKPAMIASASMGGFVGLLAAFMPNLLAFSICKFCGSLFLSSASSLGFAYICEMLPVKSRDRTVLICNGLVMILSTLCPGKLPPAFFLLPL
Summary
Uniprot
A0A212EWS0
A0A2A4JMQ4
A0A2A4JM76
A0A2H1WNU1
A0A0N1IHI8
A0A194Q1N6
+ More
A0A194QTS6 A0A212EYD8 A0A2A4K5Q7 A0A194QN48 A0A194PP30 A0A194QN61 A0A194PJI3 A0A2H1WYT6 A0A2H1X0I9 A0A194Q1R5 H9JLM1 A0A212EY03 A0A194Q2D2 A0A2A4JKW1 A0A2H1W7K7 A0A0L7L0V3 A0A2A4JLN1 A0A2A4JX40 H9JVR6 A0A1B6KCI1 A0A212EZN8 A0A1Q3FRJ7 A0A0N1IPG1 A0A1B6I2U3 A0A1B6LQJ0 A0A1B6KRD4 A0A1B6G3M6 D6WB99 A0A2H1WYY1 A0A034VLH4 W8BIF4 A0A1B6MGQ6 E0VS92 A0A1B6KBY7 A0A1B6KRY4 W8C469 A0A1B6EYA3 A0A1B6KVL9 A0A0K8TPB6 A0A2A4JL38 A0A0K8UH50 A0A0N1IG89 A0A3L8DJU1 A0A2H1VMI9 W8BDL8 H9IW67 A0A026WFC5 K7IYL5 A0A1B0CB10 Q16LN9 A0A336MF81 A0A1B0CYW8 A0A1B0GJR0 A0A1B6EF96 A0A2H1W801 D6W821 A0A1Y1L833 A0A212ESA5 A0A212ESJ6 E2B477 A0A1B0CZL0 A0A158NM89 A0A1B0CG42 A0A0A1XE46 A0A0K8SJZ1 B0WDR9 B3P9C6 A0A139WMM0 S4PYD7 B0WKE6 U5EQ91 A0A146LC37 Q16F34 A0A194RSD6 A0A1S4G380 A0A0A9WZX1 A0A212EQC0 A0A0N1I5D8 A0A2H1WI98 A0A0A1XKB5 A0A2J7RSM1 V5GNJ1 A0A182SX71 A0A0L0BTJ0 A0A336LV65 A0A1L8DV70 A0A2H1W1V2 A0A182M591 A0A2A4J951 A0A182KFV8
A0A194QTS6 A0A212EYD8 A0A2A4K5Q7 A0A194QN48 A0A194PP30 A0A194QN61 A0A194PJI3 A0A2H1WYT6 A0A2H1X0I9 A0A194Q1R5 H9JLM1 A0A212EY03 A0A194Q2D2 A0A2A4JKW1 A0A2H1W7K7 A0A0L7L0V3 A0A2A4JLN1 A0A2A4JX40 H9JVR6 A0A1B6KCI1 A0A212EZN8 A0A1Q3FRJ7 A0A0N1IPG1 A0A1B6I2U3 A0A1B6LQJ0 A0A1B6KRD4 A0A1B6G3M6 D6WB99 A0A2H1WYY1 A0A034VLH4 W8BIF4 A0A1B6MGQ6 E0VS92 A0A1B6KBY7 A0A1B6KRY4 W8C469 A0A1B6EYA3 A0A1B6KVL9 A0A0K8TPB6 A0A2A4JL38 A0A0K8UH50 A0A0N1IG89 A0A3L8DJU1 A0A2H1VMI9 W8BDL8 H9IW67 A0A026WFC5 K7IYL5 A0A1B0CB10 Q16LN9 A0A336MF81 A0A1B0CYW8 A0A1B0GJR0 A0A1B6EF96 A0A2H1W801 D6W821 A0A1Y1L833 A0A212ESA5 A0A212ESJ6 E2B477 A0A1B0CZL0 A0A158NM89 A0A1B0CG42 A0A0A1XE46 A0A0K8SJZ1 B0WDR9 B3P9C6 A0A139WMM0 S4PYD7 B0WKE6 U5EQ91 A0A146LC37 Q16F34 A0A194RSD6 A0A1S4G380 A0A0A9WZX1 A0A212EQC0 A0A0N1I5D8 A0A2H1WI98 A0A0A1XKB5 A0A2J7RSM1 V5GNJ1 A0A182SX71 A0A0L0BTJ0 A0A336LV65 A0A1L8DV70 A0A2H1W1V2 A0A182M591 A0A2A4J951 A0A182KFV8
Pubmed
EMBL
AGBW02011907
OWR45939.1
NWSH01001089
PCG72682.1
PCG72684.1
ODYU01009931
+ More
SOQ54677.1 KQ460542 KPJ14128.1 KQ459579 KPI99492.1 KQ461194 KPJ06946.1 AGBW02011531 OWR46513.1 NWSH01000098 PCG79585.1 KPJ06948.1 KQ459603 KPI92885.1 KPJ06947.1 KPI92884.1 ODYU01012108 SOQ58229.1 ODYU01012422 SOQ58716.1 KPI99491.1 BABH01005452 AGBW02011653 OWR46341.1 KPI99493.1 PCG72687.1 ODYU01006861 SOQ49085.1 JTDY01003749 KOB69050.1 PCG72686.1 NWSH01000457 PCG76268.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 GEBQ01030862 JAT09115.1 AGBW02011241 OWR46942.1 GFDL01004992 JAV30053.1 KPJ14127.1 GECU01026455 JAS81251.1 GEBQ01014128 JAT25849.1 GEBQ01025979 JAT13998.1 GECZ01012721 JAS57048.1 KQ971312 EEZ97916.1 ODYU01012071 SOQ58176.1 GAKP01015980 JAC42972.1 GAMC01009767 JAB96788.1 GEBQ01004874 JAT35103.1 DS235746 EEB16248.1 GEBQ01031010 JAT08967.1 GEBQ01025779 JAT14198.1 GAMC01009766 JAB96789.1 GECZ01027136 JAS42633.1 GEBQ01024546 JAT15431.1 GDAI01001409 JAI16194.1 PCG72681.1 GDHF01026325 JAI25989.1 KPJ14129.1 QOIP01000007 RLU20667.1 ODYU01003368 SOQ42041.1 GAMC01009768 JAB96787.1 BABH01005928 BABH01005929 BABH01005930 KK107238 EZA54618.1 AJWK01004723 CH477898 EAT35249.1 UFQT01000487 SSX24708.1 AJVK01020464 AJVK01020465 AJVK01020466 AJWK01022122 AJWK01022123 GEDC01000686 JAS36612.1 SOQ49086.1 KQ971307 EFA11011.1 GEZM01068064 GEZM01068063 JAV67237.1 AGBW02012834 OWR44382.1 AGBW02012778 OWR44449.1 GL445515 EFN89494.1 AJVK01009629 AJVK01009630 ADTU01020150 AJWK01010713 AJWK01010714 AJWK01010715 GBXI01004603 JAD09689.1 GBRD01012207 JAG53617.1 DS231900 EDS44822.1 CH954183 EDV45422.2 KYB29087.1 GAIX01003468 JAA89092.1 DS231971 EDS29764.1 GANO01004357 JAB55514.1 GDHC01014499 JAQ04130.1 CH478516 EAT32850.1 KQ459984 KPJ18981.1 GBHO01030315 JAG13289.1 AGBW02013272 OWR43679.1 KQ461068 KPJ09887.1 ODYU01008516 SOQ52184.1 GBXI01002836 JAD11456.1 NEVH01000252 PNF43835.1 GALX01002772 JAB65694.1 JRES01001365 KNC23336.1 UFQT01000207 SSX21730.1 GFDF01003899 JAV10185.1 ODYU01005784 SOQ47007.1 AXCM01004479 NWSH01002480 PCG68208.1
SOQ54677.1 KQ460542 KPJ14128.1 KQ459579 KPI99492.1 KQ461194 KPJ06946.1 AGBW02011531 OWR46513.1 NWSH01000098 PCG79585.1 KPJ06948.1 KQ459603 KPI92885.1 KPJ06947.1 KPI92884.1 ODYU01012108 SOQ58229.1 ODYU01012422 SOQ58716.1 KPI99491.1 BABH01005452 AGBW02011653 OWR46341.1 KPI99493.1 PCG72687.1 ODYU01006861 SOQ49085.1 JTDY01003749 KOB69050.1 PCG72686.1 NWSH01000457 PCG76268.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 GEBQ01030862 JAT09115.1 AGBW02011241 OWR46942.1 GFDL01004992 JAV30053.1 KPJ14127.1 GECU01026455 JAS81251.1 GEBQ01014128 JAT25849.1 GEBQ01025979 JAT13998.1 GECZ01012721 JAS57048.1 KQ971312 EEZ97916.1 ODYU01012071 SOQ58176.1 GAKP01015980 JAC42972.1 GAMC01009767 JAB96788.1 GEBQ01004874 JAT35103.1 DS235746 EEB16248.1 GEBQ01031010 JAT08967.1 GEBQ01025779 JAT14198.1 GAMC01009766 JAB96789.1 GECZ01027136 JAS42633.1 GEBQ01024546 JAT15431.1 GDAI01001409 JAI16194.1 PCG72681.1 GDHF01026325 JAI25989.1 KPJ14129.1 QOIP01000007 RLU20667.1 ODYU01003368 SOQ42041.1 GAMC01009768 JAB96787.1 BABH01005928 BABH01005929 BABH01005930 KK107238 EZA54618.1 AJWK01004723 CH477898 EAT35249.1 UFQT01000487 SSX24708.1 AJVK01020464 AJVK01020465 AJVK01020466 AJWK01022122 AJWK01022123 GEDC01000686 JAS36612.1 SOQ49086.1 KQ971307 EFA11011.1 GEZM01068064 GEZM01068063 JAV67237.1 AGBW02012834 OWR44382.1 AGBW02012778 OWR44449.1 GL445515 EFN89494.1 AJVK01009629 AJVK01009630 ADTU01020150 AJWK01010713 AJWK01010714 AJWK01010715 GBXI01004603 JAD09689.1 GBRD01012207 JAG53617.1 DS231900 EDS44822.1 CH954183 EDV45422.2 KYB29087.1 GAIX01003468 JAA89092.1 DS231971 EDS29764.1 GANO01004357 JAB55514.1 GDHC01014499 JAQ04130.1 CH478516 EAT32850.1 KQ459984 KPJ18981.1 GBHO01030315 JAG13289.1 AGBW02013272 OWR43679.1 KQ461068 KPJ09887.1 ODYU01008516 SOQ52184.1 GBXI01002836 JAD11456.1 NEVH01000252 PNF43835.1 GALX01002772 JAB65694.1 JRES01001365 KNC23336.1 UFQT01000207 SSX21730.1 GFDF01003899 JAV10185.1 ODYU01005784 SOQ47007.1 AXCM01004479 NWSH01002480 PCG68208.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A212EWS0
A0A2A4JMQ4
A0A2A4JM76
A0A2H1WNU1
A0A0N1IHI8
A0A194Q1N6
+ More
A0A194QTS6 A0A212EYD8 A0A2A4K5Q7 A0A194QN48 A0A194PP30 A0A194QN61 A0A194PJI3 A0A2H1WYT6 A0A2H1X0I9 A0A194Q1R5 H9JLM1 A0A212EY03 A0A194Q2D2 A0A2A4JKW1 A0A2H1W7K7 A0A0L7L0V3 A0A2A4JLN1 A0A2A4JX40 H9JVR6 A0A1B6KCI1 A0A212EZN8 A0A1Q3FRJ7 A0A0N1IPG1 A0A1B6I2U3 A0A1B6LQJ0 A0A1B6KRD4 A0A1B6G3M6 D6WB99 A0A2H1WYY1 A0A034VLH4 W8BIF4 A0A1B6MGQ6 E0VS92 A0A1B6KBY7 A0A1B6KRY4 W8C469 A0A1B6EYA3 A0A1B6KVL9 A0A0K8TPB6 A0A2A4JL38 A0A0K8UH50 A0A0N1IG89 A0A3L8DJU1 A0A2H1VMI9 W8BDL8 H9IW67 A0A026WFC5 K7IYL5 A0A1B0CB10 Q16LN9 A0A336MF81 A0A1B0CYW8 A0A1B0GJR0 A0A1B6EF96 A0A2H1W801 D6W821 A0A1Y1L833 A0A212ESA5 A0A212ESJ6 E2B477 A0A1B0CZL0 A0A158NM89 A0A1B0CG42 A0A0A1XE46 A0A0K8SJZ1 B0WDR9 B3P9C6 A0A139WMM0 S4PYD7 B0WKE6 U5EQ91 A0A146LC37 Q16F34 A0A194RSD6 A0A1S4G380 A0A0A9WZX1 A0A212EQC0 A0A0N1I5D8 A0A2H1WI98 A0A0A1XKB5 A0A2J7RSM1 V5GNJ1 A0A182SX71 A0A0L0BTJ0 A0A336LV65 A0A1L8DV70 A0A2H1W1V2 A0A182M591 A0A2A4J951 A0A182KFV8
A0A194QTS6 A0A212EYD8 A0A2A4K5Q7 A0A194QN48 A0A194PP30 A0A194QN61 A0A194PJI3 A0A2H1WYT6 A0A2H1X0I9 A0A194Q1R5 H9JLM1 A0A212EY03 A0A194Q2D2 A0A2A4JKW1 A0A2H1W7K7 A0A0L7L0V3 A0A2A4JLN1 A0A2A4JX40 H9JVR6 A0A1B6KCI1 A0A212EZN8 A0A1Q3FRJ7 A0A0N1IPG1 A0A1B6I2U3 A0A1B6LQJ0 A0A1B6KRD4 A0A1B6G3M6 D6WB99 A0A2H1WYY1 A0A034VLH4 W8BIF4 A0A1B6MGQ6 E0VS92 A0A1B6KBY7 A0A1B6KRY4 W8C469 A0A1B6EYA3 A0A1B6KVL9 A0A0K8TPB6 A0A2A4JL38 A0A0K8UH50 A0A0N1IG89 A0A3L8DJU1 A0A2H1VMI9 W8BDL8 H9IW67 A0A026WFC5 K7IYL5 A0A1B0CB10 Q16LN9 A0A336MF81 A0A1B0CYW8 A0A1B0GJR0 A0A1B6EF96 A0A2H1W801 D6W821 A0A1Y1L833 A0A212ESA5 A0A212ESJ6 E2B477 A0A1B0CZL0 A0A158NM89 A0A1B0CG42 A0A0A1XE46 A0A0K8SJZ1 B0WDR9 B3P9C6 A0A139WMM0 S4PYD7 B0WKE6 U5EQ91 A0A146LC37 Q16F34 A0A194RSD6 A0A1S4G380 A0A0A9WZX1 A0A212EQC0 A0A0N1I5D8 A0A2H1WI98 A0A0A1XKB5 A0A2J7RSM1 V5GNJ1 A0A182SX71 A0A0L0BTJ0 A0A336LV65 A0A1L8DV70 A0A2H1W1V2 A0A182M591 A0A2A4J951 A0A182KFV8
Ontologies
GO
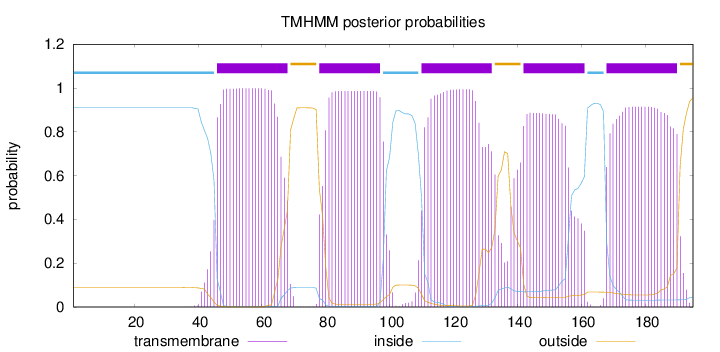
Topology
Length:
195
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
102.77764
Exp number, first 60 AAs:
15.78057
Total prob of N-in:
0.91147
POSSIBLE N-term signal
sequence
inside
1 - 45
TMhelix
46 - 68
outside
69 - 77
TMhelix
78 - 97
inside
98 - 109
TMhelix
110 - 132
outside
133 - 141
TMhelix
142 - 161
inside
162 - 167
TMhelix
168 - 190
outside
191 - 195
Population Genetic Test Statistics
Pi
231.629077
Theta
141.879639
Tajima's D
-1.742631
CLR
1365.900862
CSRT
0.0360481975901205
Interpretation
Uncertain
