Gene
KWMTBOMO03221
Annotation
PREDICTED:_synaptic_vesicle_glycoprotein_2C-like?_partial_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.865
Sequence
CDS
ATGGGTGCGATTATCTTTAGACCGTGGCGTCTTCTAACCATCGTCTACGCTATTCCATTGCTCGTAGTGGCGGTCTGCATGACCTGGATGCAGGAGAGCCCCAAATTCCTAGTCACGAGAGGCAAACGCACCGAAGCTCTGGCTGTCCTCAGGAAAATATACTCCATTAACAGGGGAAAGCCTGAAAAAGAATATTGTGTCAAATCATTGAGTTGTCTATCAGAAACAGCCAGCGACGCCAGTGACACTGATGAGCTCAGAAATAATTCTATACTTTCGTTGCTTCGACCGCCGCATCTGAAATGGCTGCTTTTAACAGGATTCCTCATGTTTGGACTGTTCTCATTATTGAATGGTCTCTTTCTATTCGCACCGGACACGATCAATAGAATGCTGACTGGAACATCAGACGAAGTCGGAACCATATGTCTGTATATGAACAACGGTAACAATATAACGGCGATTACTGACGCGTGCGTGGATACGATCTCAAACAGCACCCTCCTAGTGATGGTGGTCACGACACTTGTGTACAGCGTCATTGTGATGGCGATAAGTGTCGCCCCACTGTCCAAGAAGACTCTGCTGGTCGCCGTATTCTTTGTGGTGGGCATCAGCTGTCTCGTTTCTGGCCTCAGCAAGAACAGAATGCTGGCCGGTGTTGCCATGTCGTCATTACAAATAACTGCTCTGGGTATCGGACCTTTGACTGCCTATGCGATACATCTGTTTCCAACGAGCTTAAGAGGAACTGCAGTAGGAGCCGTGATGATGTTTGGCAGGATCGGATCCGCGTTAGGCGCGAATGCAGCTGGATTCTTCCTATCGATAGCTTGTACCCTTACCTTCTATGGATTCTCGTTCATGCTGTTTTTGTGTGCTGCCCTCAGTTTGCTGCTGCCAAAAGACCGAAGTTGA
Protein
MGAIIFRPWRLLTIVYAIPLLVVAVCMTWMQESPKFLVTRGKRTEALAVLRKIYSINRGKPEKEYCVKSLSCLSETASDASDTDELRNNSILSLLRPPHLKWLLLTGFLMFGLFSLLNGLFLFAPDTINRMLTGTSDEVGTICLYMNNGNNITAITDACVDTISNSTLLVMVVTTLVYSVIVMAISVAPLSKKTLLVAVFFVVGISCLVSGLSKNRMLAGVAMSSLQITALGIGPLTAYAIHLFPTSLRGTAVGAVMMFGRIGSALGANAAGFFLSIACTLTFYGFSFMLFLCAALSLLLPKDRS
Summary
Uniprot
A0A2A4JM76
A0A2A4JMQ4
A0A2H1WNU1
A0A194Q1N6
A0A212EWS0
A0A0N1IHI8
+ More
A0A2A4K5Q7 A0A2H1X0I9 A0A2W1BLZ7 A0A2H1WYT6 A0A212EYD8 A0A194QN61 A0A194PP30 A0A194QN48 A0A194PJI3 A0A212EZN8 D6WB98 A0A2H1W7K7 A0A2A4JKW1 A0A2A4JLN1 A0A2H1WJU4 A0A2H1WI98 A0A2W1BUU8 A0A1B6MNI5 H9JVR6 A0A0N1IG89 A0A194Q1T1 A0A2A4JL38 A0A194Q2D2 A0A182VKS7 A0A2W1BGE8 H9JVL1 A0A182INB7 N6T4U4 A0A2A4IYX6 Q6U1H2 A0A194Q1T6 A0A2H1V250 T1GJI8 A0A182WUC2 A0A182IB08 F5HLT0 A0A1B6BZJ7 B0WQD3 A0A182I9F6 B0XE98 A0A0L7L0V3 A0A1B0D3C4 A0A2H1WUV1 A0A2H1WGI5 A0A336LUV1 T1GJ80 A0A1B6KVL9 A0A2H1VUC9 A0A0M5J530 A0A2W1BVQ5 A0A212F5C2 A0A182IRQ8
A0A2A4K5Q7 A0A2H1X0I9 A0A2W1BLZ7 A0A2H1WYT6 A0A212EYD8 A0A194QN61 A0A194PP30 A0A194QN48 A0A194PJI3 A0A212EZN8 D6WB98 A0A2H1W7K7 A0A2A4JKW1 A0A2A4JLN1 A0A2H1WJU4 A0A2H1WI98 A0A2W1BUU8 A0A1B6MNI5 H9JVR6 A0A0N1IG89 A0A194Q1T1 A0A2A4JL38 A0A194Q2D2 A0A182VKS7 A0A2W1BGE8 H9JVL1 A0A182INB7 N6T4U4 A0A2A4IYX6 Q6U1H2 A0A194Q1T6 A0A2H1V250 T1GJI8 A0A182WUC2 A0A182IB08 F5HLT0 A0A1B6BZJ7 B0WQD3 A0A182I9F6 B0XE98 A0A0L7L0V3 A0A1B0D3C4 A0A2H1WUV1 A0A2H1WGI5 A0A336LUV1 T1GJ80 A0A1B6KVL9 A0A2H1VUC9 A0A0M5J530 A0A2W1BVQ5 A0A212F5C2 A0A182IRQ8
Pubmed
EMBL
NWSH01001089
PCG72684.1
PCG72682.1
ODYU01009931
SOQ54677.1
KQ459579
+ More
KPI99492.1 AGBW02011907 OWR45939.1 KQ460542 KPJ14128.1 NWSH01000098 PCG79585.1 ODYU01012422 SOQ58716.1 KZ149975 PZC75948.1 ODYU01012108 SOQ58229.1 AGBW02011531 OWR46513.1 KQ461194 KPJ06947.1 KQ459603 KPI92885.1 KPJ06948.1 KPI92884.1 AGBW02011241 OWR46942.1 KQ971312 EEZ97917.1 ODYU01006861 SOQ49085.1 PCG72687.1 PCG72686.1 ODYU01009141 SOQ53353.1 ODYU01008516 SOQ52184.1 KZ149911 PZC78141.1 GEBQ01002460 JAT37517.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 KPJ14129.1 KPI99506.1 PCG72681.1 KPI99493.1 KZ150202 PZC72277.1 BABH01032458 APGK01052662 KB741213 ENN72643.1 NWSH01005206 PCG64352.1 AY380837 AAQ88394.1 KPI99511.1 ODYU01000151 SOQ34432.1 CAQQ02199956 APCN01006593 AAAB01008952 EGK97241.1 GEDC01030774 JAS06524.1 DS232039 EDS32804.1 APCN01000977 DS232813 EDS25872.1 JTDY01003749 KOB69050.1 AJVK01023425 ODYU01011202 SOQ56787.1 SOQ52183.1 UFQT01000141 SSX20771.1 CAQQ02394259 GEBQ01024546 JAT15431.1 ODYU01004504 SOQ44451.1 CP012526 ALC46844.1 KZ149979 PZC75843.1 AGBW02010212 OWR48924.1
KPI99492.1 AGBW02011907 OWR45939.1 KQ460542 KPJ14128.1 NWSH01000098 PCG79585.1 ODYU01012422 SOQ58716.1 KZ149975 PZC75948.1 ODYU01012108 SOQ58229.1 AGBW02011531 OWR46513.1 KQ461194 KPJ06947.1 KQ459603 KPI92885.1 KPJ06948.1 KPI92884.1 AGBW02011241 OWR46942.1 KQ971312 EEZ97917.1 ODYU01006861 SOQ49085.1 PCG72687.1 PCG72686.1 ODYU01009141 SOQ53353.1 ODYU01008516 SOQ52184.1 KZ149911 PZC78141.1 GEBQ01002460 JAT37517.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 KPJ14129.1 KPI99506.1 PCG72681.1 KPI99493.1 KZ150202 PZC72277.1 BABH01032458 APGK01052662 KB741213 ENN72643.1 NWSH01005206 PCG64352.1 AY380837 AAQ88394.1 KPI99511.1 ODYU01000151 SOQ34432.1 CAQQ02199956 APCN01006593 AAAB01008952 EGK97241.1 GEDC01030774 JAS06524.1 DS232039 EDS32804.1 APCN01000977 DS232813 EDS25872.1 JTDY01003749 KOB69050.1 AJVK01023425 ODYU01011202 SOQ56787.1 SOQ52183.1 UFQT01000141 SSX20771.1 CAQQ02394259 GEBQ01024546 JAT15431.1 ODYU01004504 SOQ44451.1 CP012526 ALC46844.1 KZ149979 PZC75843.1 AGBW02010212 OWR48924.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4JM76
A0A2A4JMQ4
A0A2H1WNU1
A0A194Q1N6
A0A212EWS0
A0A0N1IHI8
+ More
A0A2A4K5Q7 A0A2H1X0I9 A0A2W1BLZ7 A0A2H1WYT6 A0A212EYD8 A0A194QN61 A0A194PP30 A0A194QN48 A0A194PJI3 A0A212EZN8 D6WB98 A0A2H1W7K7 A0A2A4JKW1 A0A2A4JLN1 A0A2H1WJU4 A0A2H1WI98 A0A2W1BUU8 A0A1B6MNI5 H9JVR6 A0A0N1IG89 A0A194Q1T1 A0A2A4JL38 A0A194Q2D2 A0A182VKS7 A0A2W1BGE8 H9JVL1 A0A182INB7 N6T4U4 A0A2A4IYX6 Q6U1H2 A0A194Q1T6 A0A2H1V250 T1GJI8 A0A182WUC2 A0A182IB08 F5HLT0 A0A1B6BZJ7 B0WQD3 A0A182I9F6 B0XE98 A0A0L7L0V3 A0A1B0D3C4 A0A2H1WUV1 A0A2H1WGI5 A0A336LUV1 T1GJ80 A0A1B6KVL9 A0A2H1VUC9 A0A0M5J530 A0A2W1BVQ5 A0A212F5C2 A0A182IRQ8
A0A2A4K5Q7 A0A2H1X0I9 A0A2W1BLZ7 A0A2H1WYT6 A0A212EYD8 A0A194QN61 A0A194PP30 A0A194QN48 A0A194PJI3 A0A212EZN8 D6WB98 A0A2H1W7K7 A0A2A4JKW1 A0A2A4JLN1 A0A2H1WJU4 A0A2H1WI98 A0A2W1BUU8 A0A1B6MNI5 H9JVR6 A0A0N1IG89 A0A194Q1T1 A0A2A4JL38 A0A194Q2D2 A0A182VKS7 A0A2W1BGE8 H9JVL1 A0A182INB7 N6T4U4 A0A2A4IYX6 Q6U1H2 A0A194Q1T6 A0A2H1V250 T1GJI8 A0A182WUC2 A0A182IB08 F5HLT0 A0A1B6BZJ7 B0WQD3 A0A182I9F6 B0XE98 A0A0L7L0V3 A0A1B0D3C4 A0A2H1WUV1 A0A2H1WGI5 A0A336LUV1 T1GJ80 A0A1B6KVL9 A0A2H1VUC9 A0A0M5J530 A0A2W1BVQ5 A0A212F5C2 A0A182IRQ8
PDB
6N3I
E-value=0.0307352,
Score=86
Ontologies
GO
Topology
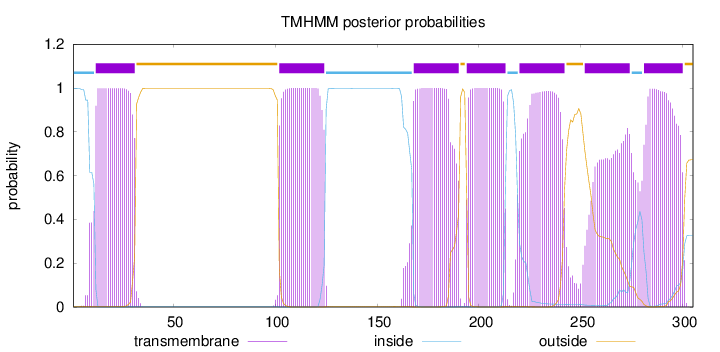
Length:
305
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
144.69738
Exp number, first 60 AAs:
20.96509
Total prob of N-in:
0.99768
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 31
outside
32 - 101
TMhelix
102 - 124
inside
125 - 167
TMhelix
168 - 190
outside
191 - 193
TMhelix
194 - 213
inside
214 - 219
TMhelix
220 - 242
outside
243 - 251
TMhelix
252 - 274
inside
275 - 280
TMhelix
281 - 300
outside
301 - 305
Population Genetic Test Statistics
Pi
164.180059
Theta
143.124392
Tajima's D
0.490949
CLR
0.850404
CSRT
0.508974551272436
Interpretation
Uncertain
