Gene
KWMTBOMO03218 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013631
Annotation
PREDICTED:_UDP-galactose_4-epimerase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.847
Sequence
CDS
ATGTCTGATACAACCGGTGATAATTTAAAACCACGAGTTTTAATACTAGGAGGATGCGGCTTCATCGGGCGGAACTTGGTAGATTACTTGATAAGGAACGATCTGGTTAGCGGGCTTCGTGTTGTGGACAAGACCCCGCCTCAACTGGCTTTCCTGAACCCTACACATTCTAAAACATTTGAGGACCCCCGCGTTGAATACAAAAGCGCAAATCTAATTAATCAGACTTCTTGCGCATCAGCGTTAGATCCAGGAGACGACGCGCCATGGGGGCTGGTAGTGAATTGTGCGAGCGAAACTCGCGGAGGTCAGACCGAGGCGGTTTATGCGGAAGGAATCGTCACCCTCAGCCTGAACGTGGCCAAGCACTGCGCCAGGATGAAGGTACCAAGACTGGTAGAGATCTCAAGTGGACAGATGTGCAGCAATGATAAGCCCCAAAAGGAAGATTGCTCAATTGACCCTTGGACGATCGAAGGTCGGATGAAGAGCAAAGTGGAACAAGAGCTGAAAAACATGGAGGACCTGAACTACACCATCATAAGGCCCGCTATTGTGTACGGAATCGGAGACAGAAGAAGTTTGACACCCCGTCTCCTCTACGGTGGAATTTACAAGCATTTAGGCGAAACAATGAAACTCCTTTGGACTGGCGATCTGAAAATGAACACGGTACATGTCCGCGACGTCTGTCGTGCCATCTGGACTCTCGGGACCAGTCCTCAGGCCAACAAGCAGATCTATAACTTGGTTGACGAAGGGAACAGCACTCAGGGTACATTGGCCGAGCTCGTCTCTGATATATTCAAAATCAATCATGATTACTATGGAACCGCTATTTCTACTTTGGCTAAGAACGACATAGCGTCTGTGGCGGAGGAAGCCAACGACAAGCACCTGACCGCGTGGGCGGACATCTGCCGCAAGTACTCCCTCCAGCACACGCCGCTCGAGCCCAGCGCCGGCGTGGAGCTGCTCCTCAACAAGCAGCTCTGTCTCGACGGGACCAAGCTGCGCGCGCTGATGCAACTGGACGTGCCCAAACCTACCGCCGAACTGTTCAGAGAGGTTTTAGAAGACTATGCTTCGATGCAACTCTTCCCCAAGGAACTTTTACTATGA
Protein
MSDTTGDNLKPRVLILGGCGFIGRNLVDYLIRNDLVSGLRVVDKTPPQLAFLNPTHSKTFEDPRVEYKSANLINQTSCASALDPGDDAPWGLVVNCASETRGGQTEAVYAEGIVTLSLNVAKHCARMKVPRLVEISSGQMCSNDKPQKEDCSIDPWTIEGRMKSKVEQELKNMEDLNYTIIRPAIVYGIGDRRSLTPRLLYGGIYKHLGETMKLLWTGDLKMNTVHVRDVCRAIWTLGTSPQANKQIYNLVDEGNSTQGTLAELVSDIFKINHDYYGTAISTLAKNDIASVAEEANDKHLTAWADICRKYSLQHTPLEPSAGVELLLNKQLCLDGTKLRALMQLDVPKPTAELFREVLEDYASMQLFPKELLL
Summary
Similarity
Belongs to the 3-beta-HSD family.
Uniprot
H9JVR8
Q2F5U7
A0A212EQB7
A0A194Q1S1
A0A2H1W3G6
A0A2A4J1B7
+ More
E0VD37 A0A0L7L0C4 K7J2J2 A0A0N0PCX3 A0A2J7RRT9 A0A067RJ95 A0A2A3ED06 A0A232FFA2 A0A088ANF9 A0A154PAW1 A0A0L7RH04 F4WZW9 E2ACB9 A0A026X1D3 A0A0M8ZND2 A0A195DXJ8 A0A151XAF6 A0A0J7KDQ3 D6WAY2 E2BAR9 A0A195B727 A0A151ICG4 A0A3L8DRP1 A0A1S4FQC6 A0A087TEH3 A0A1A9ZSV9 A0A1A9VDP6 Q16SV9 A0A1A9Y0I8 A0A0V0GBH3 A0A1B6DYC1 A0A1Q3F9G2 V5GVS9 A0A1Y1MDH2 E9ILA8 B0X5C7 A0A034WSA4 A0A1A9W5Q6 W8CAV4 A0A023F1S8 A0A0A1X5U0 A0A1B6M4V8 A0A0C9QMM5 B3M318 A0A0L0BR88 A0A0K8VDX5 A0A182UAF5 A0A1I8NEJ7 A0A2M4BQW7 A0A1W7R8D1 A0A182ID31 Q7PQ88 A0A1L8EC87 A0A182L6W3 B4HG09 A0A182X8U9 A0A0K8RDN9 A0A2M4BQW2 B4QSD3 B3P8B8 Q9VCF8 A0A131XY58 A0A1I8Q2J4 L7M5Y5 A0A182V4I3 B4K739 B4PL49 A0A224YIV9 A0A182F9K5 A0A182N4Q2 A0A1W4UJT4 A0A195FF94 B7P565 A0A131YMG1 W5JG14 A0A3B0JKL6 A0A182QJ94 A0A2M4AN79 A0A182JPD5 Q298M8 B4G4C2 A0A0M5J0K5 A0A084VEN8 B4M0C6 A0A2P8XZ44 A0A131XJC8 A0A182VY51 A0A182PRH4 A0A293N0S7 A0A1E1XVL9 A0A182SE42 A0A182XWQ8 A0A182IRA6
E0VD37 A0A0L7L0C4 K7J2J2 A0A0N0PCX3 A0A2J7RRT9 A0A067RJ95 A0A2A3ED06 A0A232FFA2 A0A088ANF9 A0A154PAW1 A0A0L7RH04 F4WZW9 E2ACB9 A0A026X1D3 A0A0M8ZND2 A0A195DXJ8 A0A151XAF6 A0A0J7KDQ3 D6WAY2 E2BAR9 A0A195B727 A0A151ICG4 A0A3L8DRP1 A0A1S4FQC6 A0A087TEH3 A0A1A9ZSV9 A0A1A9VDP6 Q16SV9 A0A1A9Y0I8 A0A0V0GBH3 A0A1B6DYC1 A0A1Q3F9G2 V5GVS9 A0A1Y1MDH2 E9ILA8 B0X5C7 A0A034WSA4 A0A1A9W5Q6 W8CAV4 A0A023F1S8 A0A0A1X5U0 A0A1B6M4V8 A0A0C9QMM5 B3M318 A0A0L0BR88 A0A0K8VDX5 A0A182UAF5 A0A1I8NEJ7 A0A2M4BQW7 A0A1W7R8D1 A0A182ID31 Q7PQ88 A0A1L8EC87 A0A182L6W3 B4HG09 A0A182X8U9 A0A0K8RDN9 A0A2M4BQW2 B4QSD3 B3P8B8 Q9VCF8 A0A131XY58 A0A1I8Q2J4 L7M5Y5 A0A182V4I3 B4K739 B4PL49 A0A224YIV9 A0A182F9K5 A0A182N4Q2 A0A1W4UJT4 A0A195FF94 B7P565 A0A131YMG1 W5JG14 A0A3B0JKL6 A0A182QJ94 A0A2M4AN79 A0A182JPD5 Q298M8 B4G4C2 A0A0M5J0K5 A0A084VEN8 B4M0C6 A0A2P8XZ44 A0A131XJC8 A0A182VY51 A0A182PRH4 A0A293N0S7 A0A1E1XVL9 A0A182SE42 A0A182XWQ8 A0A182IRA6
Pubmed
19121390
22118469
26354079
20566863
26227816
20075255
+ More
24845553 28648823 21719571 20798317 24508170 18362917 19820115 30249741 17510324 28004739 21282665 25348373 24495485 25474469 25830018 17994087 26108605 25315136 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25576852 17550304 28797301 26830274 20920257 23761445 15632085 24438588 29403074 28049606 29209593 25244985
24845553 28648823 21719571 20798317 24508170 18362917 19820115 30249741 17510324 28004739 21282665 25348373 24495485 25474469 25830018 17994087 26108605 25315136 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25576852 17550304 28797301 26830274 20920257 23761445 15632085 24438588 29403074 28049606 29209593 25244985
EMBL
BABH01032430
BABH01032431
BABH01032432
BABH01032433
DQ311326
ABD36270.1
+ More
AGBW02013272 OWR43678.1 KQ459579 KPI99496.1 ODYU01006064 SOQ47588.1 NWSH01004378 PCG65173.1 DS235070 EEB11293.1 JTDY01003969 KOB68761.1 KQ460542 KPJ14132.1 NEVH01000598 PNF43558.1 KK852651 KDR19426.1 KZ288292 PBC29172.1 NNAY01000289 OXU29446.1 KQ434864 KZC08992.1 KQ414596 KOC69976.1 GL888480 EGI60259.1 GL438491 EFN68909.1 KK107063 EZA61184.1 KQ436008 KOX67607.1 KQ980107 KYN17625.1 KQ982339 KYQ57344.1 LBMM01009014 KMQ88462.1 KQ971312 EEZ97930.1 GL446831 EFN87204.1 KQ976574 KYM80301.1 KQ978053 KYM97777.1 QOIP01000005 RLU22843.1 KK114856 KFM63512.1 CH477664 EAT37560.1 GECL01000896 JAP05228.1 GEDC01006637 JAS30661.1 GFDL01010845 JAV24200.1 GALX01004118 JAB64348.1 GEZM01034203 JAV83869.1 GL764074 EFZ18609.1 DS232374 EDS40812.1 GAKP01001882 JAC57070.1 GAMC01001014 JAC05542.1 GBBI01003245 JAC15467.1 GBXI01008056 JAD06236.1 GEBQ01009018 JAT30959.1 GBYB01004759 JAG74526.1 CH902617 EDV42418.1 JRES01001482 KNC22531.1 GDHF01015231 JAI37083.1 GGFJ01006335 MBW55476.1 GEHC01000241 JAV47404.1 APCN01004984 AAAB01008898 EAA09169.4 GFDG01002448 JAV16351.1 CH480815 EDW43402.1 GADI01004627 JAA69181.1 GGFJ01006336 MBW55477.1 CM000364 EDX14132.1 CH954182 EDV53942.1 AE014297 BT004487 AAF56208.1 AAO42651.1 GEFM01004746 JAP71050.1 GACK01005564 JAA59470.1 CH933806 EDW16352.1 CM000160 EDW99034.1 GFPF01006430 MAA17576.1 KQ981625 KYN39058.1 ABJB010093497 ABJB010151749 ABJB010383793 ABJB010478031 ABJB010901382 DS639501 EEC01737.1 GEDV01008927 JAP79630.1 ADMH02001608 ETN61820.1 OUUW01000007 SPP82765.1 AXCN02001188 GGFK01008913 MBW42234.1 CM000070 EAL27927.1 CH479179 EDW24470.1 CP012526 ALC47894.1 ATLV01012285 KE524778 KFB36432.1 CH940650 EDW67288.1 PYGN01001136 PSN37282.1 GEFH01002129 JAP66452.1 GFWV01021053 MAA45781.1 GFAA01000248 JAU03187.1
AGBW02013272 OWR43678.1 KQ459579 KPI99496.1 ODYU01006064 SOQ47588.1 NWSH01004378 PCG65173.1 DS235070 EEB11293.1 JTDY01003969 KOB68761.1 KQ460542 KPJ14132.1 NEVH01000598 PNF43558.1 KK852651 KDR19426.1 KZ288292 PBC29172.1 NNAY01000289 OXU29446.1 KQ434864 KZC08992.1 KQ414596 KOC69976.1 GL888480 EGI60259.1 GL438491 EFN68909.1 KK107063 EZA61184.1 KQ436008 KOX67607.1 KQ980107 KYN17625.1 KQ982339 KYQ57344.1 LBMM01009014 KMQ88462.1 KQ971312 EEZ97930.1 GL446831 EFN87204.1 KQ976574 KYM80301.1 KQ978053 KYM97777.1 QOIP01000005 RLU22843.1 KK114856 KFM63512.1 CH477664 EAT37560.1 GECL01000896 JAP05228.1 GEDC01006637 JAS30661.1 GFDL01010845 JAV24200.1 GALX01004118 JAB64348.1 GEZM01034203 JAV83869.1 GL764074 EFZ18609.1 DS232374 EDS40812.1 GAKP01001882 JAC57070.1 GAMC01001014 JAC05542.1 GBBI01003245 JAC15467.1 GBXI01008056 JAD06236.1 GEBQ01009018 JAT30959.1 GBYB01004759 JAG74526.1 CH902617 EDV42418.1 JRES01001482 KNC22531.1 GDHF01015231 JAI37083.1 GGFJ01006335 MBW55476.1 GEHC01000241 JAV47404.1 APCN01004984 AAAB01008898 EAA09169.4 GFDG01002448 JAV16351.1 CH480815 EDW43402.1 GADI01004627 JAA69181.1 GGFJ01006336 MBW55477.1 CM000364 EDX14132.1 CH954182 EDV53942.1 AE014297 BT004487 AAF56208.1 AAO42651.1 GEFM01004746 JAP71050.1 GACK01005564 JAA59470.1 CH933806 EDW16352.1 CM000160 EDW99034.1 GFPF01006430 MAA17576.1 KQ981625 KYN39058.1 ABJB010093497 ABJB010151749 ABJB010383793 ABJB010478031 ABJB010901382 DS639501 EEC01737.1 GEDV01008927 JAP79630.1 ADMH02001608 ETN61820.1 OUUW01000007 SPP82765.1 AXCN02001188 GGFK01008913 MBW42234.1 CM000070 EAL27927.1 CH479179 EDW24470.1 CP012526 ALC47894.1 ATLV01012285 KE524778 KFB36432.1 CH940650 EDW67288.1 PYGN01001136 PSN37282.1 GEFH01002129 JAP66452.1 GFWV01021053 MAA45781.1 GFAA01000248 JAU03187.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000009046
UP000037510
+ More
UP000002358 UP000053240 UP000235965 UP000027135 UP000242457 UP000215335 UP000005203 UP000076502 UP000053825 UP000007755 UP000000311 UP000053097 UP000053105 UP000078492 UP000075809 UP000036403 UP000007266 UP000008237 UP000078540 UP000078542 UP000279307 UP000054359 UP000092445 UP000078200 UP000008820 UP000092443 UP000002320 UP000091820 UP000007801 UP000037069 UP000075902 UP000095301 UP000075840 UP000007062 UP000075882 UP000001292 UP000076407 UP000000304 UP000008711 UP000000803 UP000095300 UP000075903 UP000009192 UP000002282 UP000069272 UP000075884 UP000192221 UP000078541 UP000001555 UP000000673 UP000268350 UP000075886 UP000075881 UP000001819 UP000008744 UP000092553 UP000030765 UP000008792 UP000245037 UP000075920 UP000075885 UP000075901 UP000076408 UP000075880
UP000002358 UP000053240 UP000235965 UP000027135 UP000242457 UP000215335 UP000005203 UP000076502 UP000053825 UP000007755 UP000000311 UP000053097 UP000053105 UP000078492 UP000075809 UP000036403 UP000007266 UP000008237 UP000078540 UP000078542 UP000279307 UP000054359 UP000092445 UP000078200 UP000008820 UP000092443 UP000002320 UP000091820 UP000007801 UP000037069 UP000075902 UP000095301 UP000075840 UP000007062 UP000075882 UP000001292 UP000076407 UP000000304 UP000008711 UP000000803 UP000095300 UP000075903 UP000009192 UP000002282 UP000069272 UP000075884 UP000192221 UP000078541 UP000001555 UP000000673 UP000268350 UP000075886 UP000075881 UP000001819 UP000008744 UP000092553 UP000030765 UP000008792 UP000245037 UP000075920 UP000075885 UP000075901 UP000076408 UP000075880
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JVR8
Q2F5U7
A0A212EQB7
A0A194Q1S1
A0A2H1W3G6
A0A2A4J1B7
+ More
E0VD37 A0A0L7L0C4 K7J2J2 A0A0N0PCX3 A0A2J7RRT9 A0A067RJ95 A0A2A3ED06 A0A232FFA2 A0A088ANF9 A0A154PAW1 A0A0L7RH04 F4WZW9 E2ACB9 A0A026X1D3 A0A0M8ZND2 A0A195DXJ8 A0A151XAF6 A0A0J7KDQ3 D6WAY2 E2BAR9 A0A195B727 A0A151ICG4 A0A3L8DRP1 A0A1S4FQC6 A0A087TEH3 A0A1A9ZSV9 A0A1A9VDP6 Q16SV9 A0A1A9Y0I8 A0A0V0GBH3 A0A1B6DYC1 A0A1Q3F9G2 V5GVS9 A0A1Y1MDH2 E9ILA8 B0X5C7 A0A034WSA4 A0A1A9W5Q6 W8CAV4 A0A023F1S8 A0A0A1X5U0 A0A1B6M4V8 A0A0C9QMM5 B3M318 A0A0L0BR88 A0A0K8VDX5 A0A182UAF5 A0A1I8NEJ7 A0A2M4BQW7 A0A1W7R8D1 A0A182ID31 Q7PQ88 A0A1L8EC87 A0A182L6W3 B4HG09 A0A182X8U9 A0A0K8RDN9 A0A2M4BQW2 B4QSD3 B3P8B8 Q9VCF8 A0A131XY58 A0A1I8Q2J4 L7M5Y5 A0A182V4I3 B4K739 B4PL49 A0A224YIV9 A0A182F9K5 A0A182N4Q2 A0A1W4UJT4 A0A195FF94 B7P565 A0A131YMG1 W5JG14 A0A3B0JKL6 A0A182QJ94 A0A2M4AN79 A0A182JPD5 Q298M8 B4G4C2 A0A0M5J0K5 A0A084VEN8 B4M0C6 A0A2P8XZ44 A0A131XJC8 A0A182VY51 A0A182PRH4 A0A293N0S7 A0A1E1XVL9 A0A182SE42 A0A182XWQ8 A0A182IRA6
E0VD37 A0A0L7L0C4 K7J2J2 A0A0N0PCX3 A0A2J7RRT9 A0A067RJ95 A0A2A3ED06 A0A232FFA2 A0A088ANF9 A0A154PAW1 A0A0L7RH04 F4WZW9 E2ACB9 A0A026X1D3 A0A0M8ZND2 A0A195DXJ8 A0A151XAF6 A0A0J7KDQ3 D6WAY2 E2BAR9 A0A195B727 A0A151ICG4 A0A3L8DRP1 A0A1S4FQC6 A0A087TEH3 A0A1A9ZSV9 A0A1A9VDP6 Q16SV9 A0A1A9Y0I8 A0A0V0GBH3 A0A1B6DYC1 A0A1Q3F9G2 V5GVS9 A0A1Y1MDH2 E9ILA8 B0X5C7 A0A034WSA4 A0A1A9W5Q6 W8CAV4 A0A023F1S8 A0A0A1X5U0 A0A1B6M4V8 A0A0C9QMM5 B3M318 A0A0L0BR88 A0A0K8VDX5 A0A182UAF5 A0A1I8NEJ7 A0A2M4BQW7 A0A1W7R8D1 A0A182ID31 Q7PQ88 A0A1L8EC87 A0A182L6W3 B4HG09 A0A182X8U9 A0A0K8RDN9 A0A2M4BQW2 B4QSD3 B3P8B8 Q9VCF8 A0A131XY58 A0A1I8Q2J4 L7M5Y5 A0A182V4I3 B4K739 B4PL49 A0A224YIV9 A0A182F9K5 A0A182N4Q2 A0A1W4UJT4 A0A195FF94 B7P565 A0A131YMG1 W5JG14 A0A3B0JKL6 A0A182QJ94 A0A2M4AN79 A0A182JPD5 Q298M8 B4G4C2 A0A0M5J0K5 A0A084VEN8 B4M0C6 A0A2P8XZ44 A0A131XJC8 A0A182VY51 A0A182PRH4 A0A293N0S7 A0A1E1XVL9 A0A182SE42 A0A182XWQ8 A0A182IRA6
Ontologies
GO
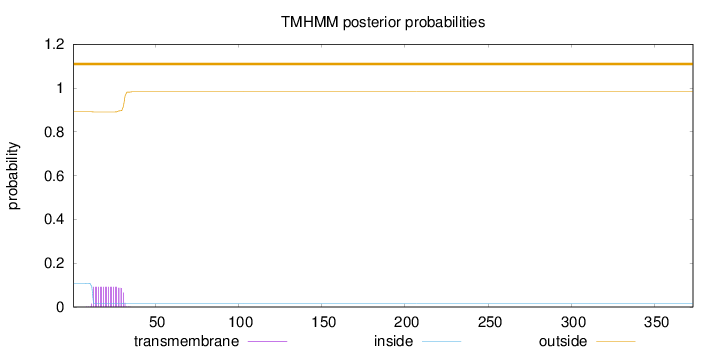
Topology
Length:
373
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.75811
Exp number, first 60 AAs:
1.75644
Total prob of N-in:
0.10838
outside
1 - 373
Population Genetic Test Statistics
Pi
185.486245
Theta
180.135553
Tajima's D
0.138425
CLR
22.556839
CSRT
0.410429478526074
Interpretation
Uncertain
