Pre Gene Modal
BGIBMGA013637
Annotation
Protein_TBRG4_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 3.52
Sequence
CDS
ATGCTAAGGTTCAGCGGATCAATATCTTGGAGGTTAGGTATCAGGGCAATCATAAGAAAGCAGTCGACTACGAACGCTGCCGCCGTAAAAACTGACAAAATCGAGGACACTAAGAATGTTCAAGGAAAAATACCCATTGCGGAAACGTTTAAGGATACTTCAGGCCTCGTAGCCGCTGCATTTGCTCACTTGAACTCTGAAAGTGAAATTACAACAACTAAGAAACCAAGTCCAAAATCAGCTAAGATAAGTCGCCACCAACAGCTCGAGAATCAAATTTCCAAAGCTGCTGATGTGAATTCTCTGCTCCTTGTTGCTGAAAATCCTGTTGTGTCTCGAAGACATGCTTTGAAGATGGTTTCTATCTTATCAGAATGGTCGAGCACTAACAAAGTGAATTTGAATGAATTCGAGAAAGACCCACGTTTTTTAAAGCTATGTAGAATTCTGGCTAGGACCTCAACTACTCATGCGATGAGTTCCTTGACAATGGCAGAGGATCTGAGCACAGTTCTAGGAATAACTGGTGATGATGAGGCAGCCCGTCTGATTTCAAATTTGACGCTGTCACAAATGATAAAGGTAATGAAGGCATTGCAGCAGAAGGGACGTCGCAGCACACCATTACTGCGTGCTCTTTCTCACAACATAACAAGTCAGGCAGAGGTCATTGATCTCAAGAAATCAGCTGATTTACTATTTTCTATGGCTAGCTTAAACTTTCCTGACCCTCTGCTACTGGACAGGATATGCAATGACGTAATAGATAGCTTACCGAAGAACGAAGACAAGCCGGCCGTAGTGAATTCGATAATTGTGTCCCTGGGCCTCATGAAGTACAGACACGAGCCGGCGCTGAACGCCATAACGGAGTGGATGATGGACCACAGCAGCGTCTGCCGCTCCAGCGACGTGGCTTCGGCCGTCGTCACGCTGGCCACCTTAAACTACGTGCCGCCCGGATCCGAAGCTCTGTTTGAGATGGCGCGCTCTCTTAAGGAGGAGGAGGTCGTGAAGCCGTCCACGTGGCTGGACTTGGTCTTCTCGCTCCTCATTCTCGACAAAGCGGATCATCATCACCTGGTATCGACTTTACGACCGGAATTCATCGATAAACTGCTCACGACTGGCGACATCCCGATCCCTTGTCGCCGGAAGCTGATGATGATAGACGCATATATCCAGCTGACGAGGCAGTCTTCCGTCCGCCTCCCCGACGACGTCCGCGAGGGCGTGCCGCTTGTCTACAACAAGGAGAAGGCGCTCTTCATTCACGCCGTGCTCGAAACATTCAGGAGTCTGGTCTCTTCTACCAACTTCCTAAAGAAGGACTGCAATTCGAACATGGGCTTCTTGTACGATGCGGAATTCGCTGTGGACGCCAAGTGTCACCCCGTGCCTCTGGACAAAGCCGCCGGTGATAAAAGCGTGTACAGGATAGCAGTGATGGCGTTGGACTACCACGACATGAGCAGAAGGACGATAGCGCCACTGGGCACCAGCGAACTGGGCGCGCGGCTGTTGCGGCTCAAGGGCTATAGAGTAATACAGATCCATTACACGGACTTCGATCCTAAAGACAAACTGGTGACGAGGGTGCAGTACATAGAGAAATTACTCAAGGAAGCTGTGAATAATAAAAACAATTAG
Protein
MLRFSGSISWRLGIRAIIRKQSTTNAAAVKTDKIEDTKNVQGKIPIAETFKDTSGLVAAAFAHLNSESEITTTKKPSPKSAKISRHQQLENQISKAADVNSLLLVAENPVVSRRHALKMVSILSEWSSTNKVNLNEFEKDPRFLKLCRILARTSTTHAMSSLTMAEDLSTVLGITGDDEAARLISNLTLSQMIKVMKALQQKGRRSTPLLRALSHNITSQAEVIDLKKSADLLFSMASLNFPDPLLLDRICNDVIDSLPKNEDKPAVVNSIIVSLGLMKYRHEPALNAITEWMMDHSSVCRSSDVASAVVTLATLNYVPPGSEALFEMARSLKEEEVVKPSTWLDLVFSLLILDKADHHHLVSTLRPEFIDKLLTTGDIPIPCRRKLMMIDAYIQLTRQSSVRLPDDVREGVPLVYNKEKALFIHAVLETFRSLVSSTNFLKKDCNSNMGFLYDAEFAVDAKCHPVPLDKAAGDKSVYRIAVMALDYHDMSRRTIAPLGTSELGARLLRLKGYRVIQIHYTDFDPKDKLVTRVQYIEKLLKEAVNNKNN
Summary
Uniprot
H9JVS4
A0A194Q3B9
A0A2H1VFA6
A0A2W1BUT8
A0A0N0PCX6
A0A2A4IZI3
+ More
A0A2A4IQY0 A0A212F2H3 U5EU15 A0A336LU09 A0A2J7QU97 A0A3B0K1M9 A0A1Y1KU67 B4LXU0 A0A2P8ZPJ1 B4JTY0 B4JRB6 A0A1B0BWH5 A0A1A9WNM3 B4G4N5 A0A1B6DEQ9 Q298T6 A0A3B0K1H5 A0A1I8MKR3 A0A1B6CQU6 B4K5D2 A0A1A9XMB2 T1PBN6 J3JXV2 N6TVI4 A0A1A9V8R6 B4NI34 D3TPV4 A0A0T6B588 A0A0M4F6B4 A0A1B0A7V6 A0A067QJ37 A0A0A1WGJ1 Q9VD14 W8B577 A0A0L0CJ33 A0A1W4W2F2 B3M224 B4PLP5 Q16U20 B0W8P8 A0A0K8WKD2 A0A1I8PS97 A0A1Q3FBW7 A0A034WBD3 A0A1S4FPI9 A0A1Q3FBE4 A0A182JBZ6 A0A084VG06 A0A146LFU5 A0A0A9WMB6 A0A146L1G7 A0A182JSE8 J9JL97 A0A182R671 D6X3J3 A0A139WB27 A0A182H8C4 T1HRH0 A0A0V0G942 A0A182QQ10 A0A182X0B3 A0A182N435 W5JF54 B4HE81 Q7PYQ7 A0A182VFZ5 A0A182I003 A0A182LR44 A0A182F972 A0A182P566 A0A182TWY5 A0A2M4BIG8 A0A023F121 A0A1B6GDU3 A0A2M4AP06 A0A1J1I3K7 A0A1B0FI20 A0A182VZ69 A0A182YAP9 A0A1B6K739 A0A2S2NBI1 B3P7J3 A0A0K8T5C1 A0A182SGH8 A0A2R7VUX8 A0A069DUQ7 A0A224XBA1 K7J0N5 A0A232F4J0 A0A182MIC2
A0A2A4IQY0 A0A212F2H3 U5EU15 A0A336LU09 A0A2J7QU97 A0A3B0K1M9 A0A1Y1KU67 B4LXU0 A0A2P8ZPJ1 B4JTY0 B4JRB6 A0A1B0BWH5 A0A1A9WNM3 B4G4N5 A0A1B6DEQ9 Q298T6 A0A3B0K1H5 A0A1I8MKR3 A0A1B6CQU6 B4K5D2 A0A1A9XMB2 T1PBN6 J3JXV2 N6TVI4 A0A1A9V8R6 B4NI34 D3TPV4 A0A0T6B588 A0A0M4F6B4 A0A1B0A7V6 A0A067QJ37 A0A0A1WGJ1 Q9VD14 W8B577 A0A0L0CJ33 A0A1W4W2F2 B3M224 B4PLP5 Q16U20 B0W8P8 A0A0K8WKD2 A0A1I8PS97 A0A1Q3FBW7 A0A034WBD3 A0A1S4FPI9 A0A1Q3FBE4 A0A182JBZ6 A0A084VG06 A0A146LFU5 A0A0A9WMB6 A0A146L1G7 A0A182JSE8 J9JL97 A0A182R671 D6X3J3 A0A139WB27 A0A182H8C4 T1HRH0 A0A0V0G942 A0A182QQ10 A0A182X0B3 A0A182N435 W5JF54 B4HE81 Q7PYQ7 A0A182VFZ5 A0A182I003 A0A182LR44 A0A182F972 A0A182P566 A0A182TWY5 A0A2M4BIG8 A0A023F121 A0A1B6GDU3 A0A2M4AP06 A0A1J1I3K7 A0A1B0FI20 A0A182VZ69 A0A182YAP9 A0A1B6K739 A0A2S2NBI1 B3P7J3 A0A0K8T5C1 A0A182SGH8 A0A2R7VUX8 A0A069DUQ7 A0A224XBA1 K7J0N5 A0A232F4J0 A0A182MIC2
Pubmed
19121390
26354079
28756777
22118469
28004739
17994087
+ More
29403074 15632085 25315136 22516182 23537049 20353571 24845553 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 26108605 17550304 17510324 25348373 24438588 26823975 25401762 18362917 19820115 26483478 20920257 23761445 12364791 14747013 17210077 20966253 25474469 25244985 26334808 20075255 28648823
29403074 15632085 25315136 22516182 23537049 20353571 24845553 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 26108605 17550304 17510324 25348373 24438588 26823975 25401762 18362917 19820115 26483478 20920257 23761445 12364791 14747013 17210077 20966253 25474469 25244985 26334808 20075255 28648823
EMBL
BABH01032447
KQ459579
KPI99504.1
ODYU01002258
SOQ39509.1
KZ149911
+ More
PZC78131.1 KQ460542 KPJ14142.1 NWSH01004491 PCG65049.1 NWSH01008790 PCG62417.1 AGBW02010745 OWR47923.1 GANO01004096 JAB55775.1 UFQS01000109 UFQT01000109 SSW99749.1 SSX20129.1 NEVH01011186 PNF32162.1 OUUW01000013 SPP88144.1 GEZM01077580 JAV63265.1 CH940650 EDW67899.1 PYGN01000002 PSN58393.1 CH916374 EDV91559.1 CH916373 EDV94306.1 JXJN01021768 CH479179 EDW24551.1 GEDC01013110 JAS24188.1 CM000070 EAL27869.2 SPP88145.1 GEDC01021486 JAS15812.1 CH933806 EDW16158.1 KA646177 AFP60806.1 BT128074 AEE63035.1 APGK01053205 KB741222 ENN72411.1 CH964272 EDW84726.2 EZ423456 ADD19732.1 LJIG01009835 KRT82285.1 CP012526 ALC47575.1 KK853452 KDR07530.1 GBXI01016153 JAC98138.1 AE014297 AY058310 AAF55989.3 AAL13539.1 GAMC01012815 GAMC01012813 JAB93742.1 JRES01000310 KNC32388.1 CH902617 EDV43348.1 CM000160 EDW97994.2 CH477634 EAT38022.1 DS231859 EDS39098.1 GDHF01028179 GDHF01012588 GDHF01000795 JAI24135.1 JAI39726.1 JAI51519.1 GFDL01010052 JAV24993.1 GAKP01006943 GAKP01006942 GAKP01006941 JAC52009.1 GFDL01010154 JAV24891.1 ATLV01012593 KE524806 KFB36900.1 GDHC01012777 JAQ05852.1 GBHO01034022 JAG09582.1 GDHC01017552 JAQ01077.1 ABLF02035222 KQ971374 EEZ97431.2 KYB25154.1 JXUM01118239 KQ566164 KXJ70352.1 ACPB03010481 GECL01001558 JAP04566.1 AXCN02000023 ADMH02001373 ETN62721.1 CH480815 EDW43178.1 AAAB01008987 EAA01053.4 APCN01002028 GGFJ01003696 MBW52837.1 GBBI01003996 JAC14716.1 GECZ01009144 JAS60625.1 GGFK01009190 MBW42511.1 CVRI01000040 CRK94919.1 CCAG010015572 GECU01000439 JAT07268.1 GGMR01001517 MBY14136.1 CH954182 EDV54154.1 GBRD01005094 JAG60727.1 KK854101 PTY11306.1 GBGD01001214 JAC87675.1 GFTR01006791 JAW09635.1 NNAY01001035 OXU25398.1 AXCM01018904
PZC78131.1 KQ460542 KPJ14142.1 NWSH01004491 PCG65049.1 NWSH01008790 PCG62417.1 AGBW02010745 OWR47923.1 GANO01004096 JAB55775.1 UFQS01000109 UFQT01000109 SSW99749.1 SSX20129.1 NEVH01011186 PNF32162.1 OUUW01000013 SPP88144.1 GEZM01077580 JAV63265.1 CH940650 EDW67899.1 PYGN01000002 PSN58393.1 CH916374 EDV91559.1 CH916373 EDV94306.1 JXJN01021768 CH479179 EDW24551.1 GEDC01013110 JAS24188.1 CM000070 EAL27869.2 SPP88145.1 GEDC01021486 JAS15812.1 CH933806 EDW16158.1 KA646177 AFP60806.1 BT128074 AEE63035.1 APGK01053205 KB741222 ENN72411.1 CH964272 EDW84726.2 EZ423456 ADD19732.1 LJIG01009835 KRT82285.1 CP012526 ALC47575.1 KK853452 KDR07530.1 GBXI01016153 JAC98138.1 AE014297 AY058310 AAF55989.3 AAL13539.1 GAMC01012815 GAMC01012813 JAB93742.1 JRES01000310 KNC32388.1 CH902617 EDV43348.1 CM000160 EDW97994.2 CH477634 EAT38022.1 DS231859 EDS39098.1 GDHF01028179 GDHF01012588 GDHF01000795 JAI24135.1 JAI39726.1 JAI51519.1 GFDL01010052 JAV24993.1 GAKP01006943 GAKP01006942 GAKP01006941 JAC52009.1 GFDL01010154 JAV24891.1 ATLV01012593 KE524806 KFB36900.1 GDHC01012777 JAQ05852.1 GBHO01034022 JAG09582.1 GDHC01017552 JAQ01077.1 ABLF02035222 KQ971374 EEZ97431.2 KYB25154.1 JXUM01118239 KQ566164 KXJ70352.1 ACPB03010481 GECL01001558 JAP04566.1 AXCN02000023 ADMH02001373 ETN62721.1 CH480815 EDW43178.1 AAAB01008987 EAA01053.4 APCN01002028 GGFJ01003696 MBW52837.1 GBBI01003996 JAC14716.1 GECZ01009144 JAS60625.1 GGFK01009190 MBW42511.1 CVRI01000040 CRK94919.1 CCAG010015572 GECU01000439 JAT07268.1 GGMR01001517 MBY14136.1 CH954182 EDV54154.1 GBRD01005094 JAG60727.1 KK854101 PTY11306.1 GBGD01001214 JAC87675.1 GFTR01006791 JAW09635.1 NNAY01001035 OXU25398.1 AXCM01018904
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000235965
+ More
UP000268350 UP000008792 UP000245037 UP000001070 UP000092460 UP000091820 UP000008744 UP000001819 UP000095301 UP000009192 UP000092443 UP000019118 UP000078200 UP000007798 UP000092553 UP000092445 UP000027135 UP000000803 UP000037069 UP000192221 UP000007801 UP000002282 UP000008820 UP000002320 UP000095300 UP000075880 UP000030765 UP000075881 UP000007819 UP000075900 UP000007266 UP000069940 UP000249989 UP000015103 UP000075886 UP000076407 UP000075884 UP000000673 UP000001292 UP000007062 UP000075903 UP000075840 UP000075882 UP000069272 UP000075885 UP000075902 UP000183832 UP000092444 UP000075920 UP000076408 UP000008711 UP000075901 UP000002358 UP000215335 UP000075883
UP000268350 UP000008792 UP000245037 UP000001070 UP000092460 UP000091820 UP000008744 UP000001819 UP000095301 UP000009192 UP000092443 UP000019118 UP000078200 UP000007798 UP000092553 UP000092445 UP000027135 UP000000803 UP000037069 UP000192221 UP000007801 UP000002282 UP000008820 UP000002320 UP000095300 UP000075880 UP000030765 UP000075881 UP000007819 UP000075900 UP000007266 UP000069940 UP000249989 UP000015103 UP000075886 UP000076407 UP000075884 UP000000673 UP000001292 UP000007062 UP000075903 UP000075840 UP000075882 UP000069272 UP000075885 UP000075902 UP000183832 UP000092444 UP000075920 UP000076408 UP000008711 UP000075901 UP000002358 UP000215335 UP000075883
Interpro
ProteinModelPortal
H9JVS4
A0A194Q3B9
A0A2H1VFA6
A0A2W1BUT8
A0A0N0PCX6
A0A2A4IZI3
+ More
A0A2A4IQY0 A0A212F2H3 U5EU15 A0A336LU09 A0A2J7QU97 A0A3B0K1M9 A0A1Y1KU67 B4LXU0 A0A2P8ZPJ1 B4JTY0 B4JRB6 A0A1B0BWH5 A0A1A9WNM3 B4G4N5 A0A1B6DEQ9 Q298T6 A0A3B0K1H5 A0A1I8MKR3 A0A1B6CQU6 B4K5D2 A0A1A9XMB2 T1PBN6 J3JXV2 N6TVI4 A0A1A9V8R6 B4NI34 D3TPV4 A0A0T6B588 A0A0M4F6B4 A0A1B0A7V6 A0A067QJ37 A0A0A1WGJ1 Q9VD14 W8B577 A0A0L0CJ33 A0A1W4W2F2 B3M224 B4PLP5 Q16U20 B0W8P8 A0A0K8WKD2 A0A1I8PS97 A0A1Q3FBW7 A0A034WBD3 A0A1S4FPI9 A0A1Q3FBE4 A0A182JBZ6 A0A084VG06 A0A146LFU5 A0A0A9WMB6 A0A146L1G7 A0A182JSE8 J9JL97 A0A182R671 D6X3J3 A0A139WB27 A0A182H8C4 T1HRH0 A0A0V0G942 A0A182QQ10 A0A182X0B3 A0A182N435 W5JF54 B4HE81 Q7PYQ7 A0A182VFZ5 A0A182I003 A0A182LR44 A0A182F972 A0A182P566 A0A182TWY5 A0A2M4BIG8 A0A023F121 A0A1B6GDU3 A0A2M4AP06 A0A1J1I3K7 A0A1B0FI20 A0A182VZ69 A0A182YAP9 A0A1B6K739 A0A2S2NBI1 B3P7J3 A0A0K8T5C1 A0A182SGH8 A0A2R7VUX8 A0A069DUQ7 A0A224XBA1 K7J0N5 A0A232F4J0 A0A182MIC2
A0A2A4IQY0 A0A212F2H3 U5EU15 A0A336LU09 A0A2J7QU97 A0A3B0K1M9 A0A1Y1KU67 B4LXU0 A0A2P8ZPJ1 B4JTY0 B4JRB6 A0A1B0BWH5 A0A1A9WNM3 B4G4N5 A0A1B6DEQ9 Q298T6 A0A3B0K1H5 A0A1I8MKR3 A0A1B6CQU6 B4K5D2 A0A1A9XMB2 T1PBN6 J3JXV2 N6TVI4 A0A1A9V8R6 B4NI34 D3TPV4 A0A0T6B588 A0A0M4F6B4 A0A1B0A7V6 A0A067QJ37 A0A0A1WGJ1 Q9VD14 W8B577 A0A0L0CJ33 A0A1W4W2F2 B3M224 B4PLP5 Q16U20 B0W8P8 A0A0K8WKD2 A0A1I8PS97 A0A1Q3FBW7 A0A034WBD3 A0A1S4FPI9 A0A1Q3FBE4 A0A182JBZ6 A0A084VG06 A0A146LFU5 A0A0A9WMB6 A0A146L1G7 A0A182JSE8 J9JL97 A0A182R671 D6X3J3 A0A139WB27 A0A182H8C4 T1HRH0 A0A0V0G942 A0A182QQ10 A0A182X0B3 A0A182N435 W5JF54 B4HE81 Q7PYQ7 A0A182VFZ5 A0A182I003 A0A182LR44 A0A182F972 A0A182P566 A0A182TWY5 A0A2M4BIG8 A0A023F121 A0A1B6GDU3 A0A2M4AP06 A0A1J1I3K7 A0A1B0FI20 A0A182VZ69 A0A182YAP9 A0A1B6K739 A0A2S2NBI1 B3P7J3 A0A0K8T5C1 A0A182SGH8 A0A2R7VUX8 A0A069DUQ7 A0A224XBA1 K7J0N5 A0A232F4J0 A0A182MIC2
Ontologies
GO
PANTHER
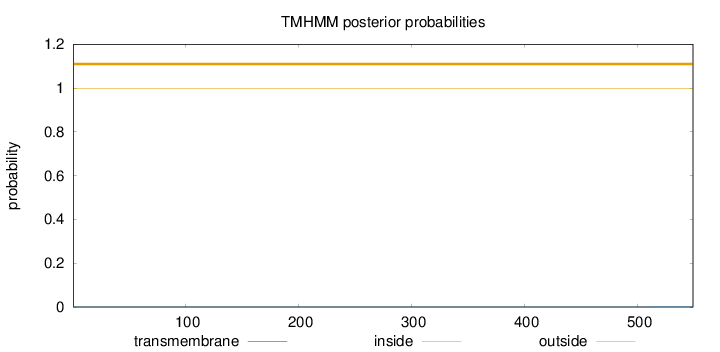
Topology
Length:
549
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00298
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00048
outside
1 - 549
Population Genetic Test Statistics
Pi
280.220614
Theta
216.309208
Tajima's D
1.133123
CLR
0.332478
CSRT
0.698515074246288
Interpretation
Uncertain
