Gene
KWMTBOMO03211
Pre Gene Modal
BGIBMGA013579
Annotation
PREDICTED:_carbonic_anhydrase_1-like_[Bombyx_mori]
Full name
Carbonic anhydrase 9
Alternative Name
Carbonate dehydratase IX
Carbonic anhydrase IX
Membrane antigen MN homolog
Carbonic anhydrase IX
Membrane antigen MN homolog
Location in the cell
Cytoplasmic Reliability : 1.133 Nuclear Reliability : 1.233
Sequence
CDS
ATGCATTTCCACTGGATTTCCGAACACTCTTTAAACGGAATTAAGTTCCCAATGGAAATACATTTCGTTCATGTTCAATCGGACCTCTCTGTCGACGAGGCTCTAATGAAGAGAGACGGCTTAGCTATCATAGCTGTGTTTTGTGAAGTACAAGCAGATTTAGACGACTACGAAAACACTCCTGTCGATGAAATAATTGAGCACCTTCCGAACCTGACGAGTACCGGAGACAGAATGAGTGGCATACTACTGGATATAAGTAAACTCCTAAGTCCTAAAAACAACAATTATTTCACGTACGCTGGATCTTTAACTTCGCCGGAGTGCAATGAGGTTGTTATTTGGATCATCTATCAATATCCTATACGCATATCGGATACGCAGCAAAAAGAATAA
Protein
MHFHWISEHSLNGIKFPMEIHFVHVQSDLSVDEALMKRDGLAIIAVFCEVQADLDDYENTPVDEIIEHLPNLTSTGDRMSGILLDISKLLSPKNNNYFTYAGSLTSPECNEVVIWIIYQYPIRISDTQQKE
Summary
Description
Reversible hydration of carbon dioxide. Participates in pH regulation (By similarity).
Catalytic Activity
H(+) + hydrogencarbonate = CO2 + H2O
Cofactor
Zn(2+)
Subunit
Forms oligomers linked by disulfide bonds.
Similarity
Belongs to the alpha-carbonic anhydrase family.
Keywords
Alternative splicing
Cell membrane
Cell projection
Complete proteome
Disulfide bond
Glycoprotein
Lyase
Membrane
Metal-binding
Phosphoprotein
Reference proteome
Signal
Transmembrane
Transmembrane helix
Zinc
Feature
chain Carbonic anhydrase 9
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JVL6
A0A212F2G6
A0A2W1BSS8
A0A2A4ISD7
A0A194Q7X0
A0A0N1IHJ4
+ More
A0A0L7L7H9 A0A2W1C1Z7 A0A2A4IRL2 A0A2H1V6M7 A0A212F719 G3TMC4 V4AH85 A0A0R3PNH9 V3ZXK9 A0A1I8CXN0 H9JVL5 A0A0L8I5X5 A0A0K0DQ44 A0A016TA97 Q3TLN7 A0A016U4Q3 A0A2P8YTE9 Q9XZG6 A0A1W4WVW2 A0A183TZ78 A0A0B2UVH4 M1EKM3 A0A2Y9DD68 A0A1U7R4S6 A0A140JWS1 A0A151N5U6 A0A1L8FCD4 T1JMA7 A0A1B0GN58 V9KKB5 E5SQL9 A0A3P4LVU6 A0A1L8F4P1 A0A016U4B9 M7BKI1 T1ISC0 F7IMS5 A0A2K6TFQ4 A0A0V1BC16 F7I0J5 A0A2K6G200 A0A0V0SHU4 A0A0V1BCN8 A0A1A8E027 A0A2R8NBR1 A0A1I8B3L8 A0A2P8XJ35 A0A091HTU2 A0A3B1JDI7 A0A2K6G203 A0A2K6FDD4 F7FL00 G1LSI9 A0A016TAD1 G3S576 Q3UUZ9 Q8VHB5 A0A3M0KBQ2 A0A3Q0D3V9 A0A3B4FV88 A0A087ZQS3 B3M1T1 A0A2Y9J8S9 A0A2K6RHV1 A0A1U7R3S6 A0A2I3G6J0 A0A3B1J929 A0A1A8PNH9 A0A2Y9E302 A0A2P2HXR5 A0A1B6GUM7 M3Y1M1 A2IBE1 A0A2U3VD21 A0A0A9X516 A0A1U7U5Z9 A0A2Y9JCI1 A0A146KNY1 A0A1D2NK15 A0A2U3YVE9 I3JGP7 A0A2R9C852 A0A2I3T6U9 A0A2J8T0E9 A0A2K6LTN4 A0A1A8RH30 L5KEW3 A0A1U7RKV7 G3SEJ4 G1SYS8 A0A3Q0GC63 A0A091GQF2
A0A0L7L7H9 A0A2W1C1Z7 A0A2A4IRL2 A0A2H1V6M7 A0A212F719 G3TMC4 V4AH85 A0A0R3PNH9 V3ZXK9 A0A1I8CXN0 H9JVL5 A0A0L8I5X5 A0A0K0DQ44 A0A016TA97 Q3TLN7 A0A016U4Q3 A0A2P8YTE9 Q9XZG6 A0A1W4WVW2 A0A183TZ78 A0A0B2UVH4 M1EKM3 A0A2Y9DD68 A0A1U7R4S6 A0A140JWS1 A0A151N5U6 A0A1L8FCD4 T1JMA7 A0A1B0GN58 V9KKB5 E5SQL9 A0A3P4LVU6 A0A1L8F4P1 A0A016U4B9 M7BKI1 T1ISC0 F7IMS5 A0A2K6TFQ4 A0A0V1BC16 F7I0J5 A0A2K6G200 A0A0V0SHU4 A0A0V1BCN8 A0A1A8E027 A0A2R8NBR1 A0A1I8B3L8 A0A2P8XJ35 A0A091HTU2 A0A3B1JDI7 A0A2K6G203 A0A2K6FDD4 F7FL00 G1LSI9 A0A016TAD1 G3S576 Q3UUZ9 Q8VHB5 A0A3M0KBQ2 A0A3Q0D3V9 A0A3B4FV88 A0A087ZQS3 B3M1T1 A0A2Y9J8S9 A0A2K6RHV1 A0A1U7R3S6 A0A2I3G6J0 A0A3B1J929 A0A1A8PNH9 A0A2Y9E302 A0A2P2HXR5 A0A1B6GUM7 M3Y1M1 A2IBE1 A0A2U3VD21 A0A0A9X516 A0A1U7U5Z9 A0A2Y9JCI1 A0A146KNY1 A0A1D2NK15 A0A2U3YVE9 I3JGP7 A0A2R9C852 A0A2I3T6U9 A0A2J8T0E9 A0A2K6LTN4 A0A1A8RH30 L5KEW3 A0A1U7RKV7 G3SEJ4 G1SYS8 A0A3Q0GC63 A0A091GQF2
EC Number
4.2.1.1
Pubmed
19121390
22118469
28756777
26354079
26227816
23254933
+ More
26829753 25730766 10349636 11042159 11076861 11217851 12466851 16141073 29403074 10222325 23236062 22293439 27762356 24402279 23624526 25243066 18809916 25329095 15057822 15632090 20010809 22398555 12040188 15489334 14604546 17994087 25362486 16831598 25401762 26823975 27289101 25186727 22722832 16136131 23258410 21993624
26829753 25730766 10349636 11042159 11076861 11217851 12466851 16141073 29403074 10222325 23236062 22293439 27762356 24402279 23624526 25243066 18809916 25329095 15057822 15632090 20010809 22398555 12040188 15489334 14604546 17994087 25362486 16831598 25401762 26823975 27289101 25186727 22722832 16136131 23258410 21993624
EMBL
BABH01032447
AGBW02010745
OWR47922.1
KZ149911
PZC78132.1
NWSH01008790
+ More
PCG62416.1 KQ459579 KPI99505.1 KQ460542 KPJ14143.1 JTDY01002414 KOB71457.1 PZC78133.1 PCG62415.1 ODYU01000955 SOQ36493.1 AGBW02009941 OWR49535.1 KB201498 ESO96282.1 UYYA01003961 VDM58193.1 ESO96283.1 KQ416468 KOF96872.1 JARK01001457 EYB99595.1 AK166403 BAE38755.1 JARK01001395 EYC09827.1 PYGN01000371 PSN47515.1 AF140537 AAD32675.1 UYWY01001209 VDM26386.1 JPKZ01003115 KHN73413.1 JP006735 AER95332.1 AB985738 BAU61548.1 AKHW03004004 KYO32204.1 CM004480 OCT69244.1 JH431506 AJVK01004379 JW865954 AFO98471.1 CYRY02002054 VCW66726.1 CM004481 OCT66537.1 EYC09826.1 KB521250 EMP37714.1 JH431429 JYDH01000063 KRY34645.1 GAMT01000477 JAB11384.1 JYDL01000008 KRX26235.1 KRY34648.1 HAEA01011057 SBQ39537.1 PYGN01001954 PSN32015.1 KL217847 KFO99718.1 AC121204 CH473962 EDL98746.1 ACTA01018962 ACTA01026962 EYB99596.1 CABD030000622 CABD030000623 BC120544 BC120546 AK136579 AK137719 CH466565 AAI20545.1 BAE23060.1 BAE23474.1 EDL02470.1 AY049077 AJ245857 AB086322 QRBI01000111 RMC10623.1 CH902617 EDV42191.2 ADFV01080394 ADFV01080395 ADFV01080396 ADFV01080397 ADFV01080398 ADFV01080399 ADFV01080400 HAEG01008708 SBR82567.1 IACF01000823 LAB66573.1 GECZ01003640 JAS66129.1 AEYP01022154 AEYP01022155 EF187254 ABM64773.1 GBHO01029697 GBHO01029696 JAG13907.1 JAG13908.1 GDHC01021234 JAP97394.1 LJIJ01000021 ODN05569.1 AERX01042727 AJFE02076650 AJFE02076651 AJFE02076652 AACZ04044244 AACZ04044245 NBAG03000330 PNI39838.1 NDHI03003533 PNJ26502.1 HAEI01008577 SBS05395.1 KB030846 ELK09033.1 AAGW02033514 KL509086 KFO85841.1
PCG62416.1 KQ459579 KPI99505.1 KQ460542 KPJ14143.1 JTDY01002414 KOB71457.1 PZC78133.1 PCG62415.1 ODYU01000955 SOQ36493.1 AGBW02009941 OWR49535.1 KB201498 ESO96282.1 UYYA01003961 VDM58193.1 ESO96283.1 KQ416468 KOF96872.1 JARK01001457 EYB99595.1 AK166403 BAE38755.1 JARK01001395 EYC09827.1 PYGN01000371 PSN47515.1 AF140537 AAD32675.1 UYWY01001209 VDM26386.1 JPKZ01003115 KHN73413.1 JP006735 AER95332.1 AB985738 BAU61548.1 AKHW03004004 KYO32204.1 CM004480 OCT69244.1 JH431506 AJVK01004379 JW865954 AFO98471.1 CYRY02002054 VCW66726.1 CM004481 OCT66537.1 EYC09826.1 KB521250 EMP37714.1 JH431429 JYDH01000063 KRY34645.1 GAMT01000477 JAB11384.1 JYDL01000008 KRX26235.1 KRY34648.1 HAEA01011057 SBQ39537.1 PYGN01001954 PSN32015.1 KL217847 KFO99718.1 AC121204 CH473962 EDL98746.1 ACTA01018962 ACTA01026962 EYB99596.1 CABD030000622 CABD030000623 BC120544 BC120546 AK136579 AK137719 CH466565 AAI20545.1 BAE23060.1 BAE23474.1 EDL02470.1 AY049077 AJ245857 AB086322 QRBI01000111 RMC10623.1 CH902617 EDV42191.2 ADFV01080394 ADFV01080395 ADFV01080396 ADFV01080397 ADFV01080398 ADFV01080399 ADFV01080400 HAEG01008708 SBR82567.1 IACF01000823 LAB66573.1 GECZ01003640 JAS66129.1 AEYP01022154 AEYP01022155 EF187254 ABM64773.1 GBHO01029697 GBHO01029696 JAG13907.1 JAG13908.1 GDHC01021234 JAP97394.1 LJIJ01000021 ODN05569.1 AERX01042727 AJFE02076650 AJFE02076651 AJFE02076652 AACZ04044244 AACZ04044245 NBAG03000330 PNI39838.1 NDHI03003533 PNJ26502.1 HAEI01008577 SBS05395.1 KB030846 ELK09033.1 AAGW02033514 KL509086 KFO85841.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
+ More
UP000007646 UP000030746 UP000050601 UP000267027 UP000095286 UP000053454 UP000035642 UP000024635 UP000245037 UP000192223 UP000050794 UP000267007 UP000031036 UP000248480 UP000189706 UP000050525 UP000186698 UP000092462 UP000031443 UP000008225 UP000233220 UP000054776 UP000233160 UP000054630 UP000095281 UP000054308 UP000018467 UP000002494 UP000008912 UP000001519 UP000000589 UP000269221 UP000261460 UP000005203 UP000007801 UP000248482 UP000233200 UP000001073 UP000000715 UP000245340 UP000189704 UP000094527 UP000245341 UP000005207 UP000240080 UP000002277 UP000233180 UP000010552 UP000189705 UP000001811
UP000007646 UP000030746 UP000050601 UP000267027 UP000095286 UP000053454 UP000035642 UP000024635 UP000245037 UP000192223 UP000050794 UP000267007 UP000031036 UP000248480 UP000189706 UP000050525 UP000186698 UP000092462 UP000031443 UP000008225 UP000233220 UP000054776 UP000233160 UP000054630 UP000095281 UP000054308 UP000018467 UP000002494 UP000008912 UP000001519 UP000000589 UP000269221 UP000261460 UP000005203 UP000007801 UP000248482 UP000233200 UP000001073 UP000000715 UP000245340 UP000189704 UP000094527 UP000245341 UP000005207 UP000240080 UP000002277 UP000233180 UP000010552 UP000189705 UP000001811
Pfam
PF00194 Carb_anhydrase
Interpro
SUPFAM
SSF51069
SSF51069
Gene 3D
ProteinModelPortal
H9JVL6
A0A212F2G6
A0A2W1BSS8
A0A2A4ISD7
A0A194Q7X0
A0A0N1IHJ4
+ More
A0A0L7L7H9 A0A2W1C1Z7 A0A2A4IRL2 A0A2H1V6M7 A0A212F719 G3TMC4 V4AH85 A0A0R3PNH9 V3ZXK9 A0A1I8CXN0 H9JVL5 A0A0L8I5X5 A0A0K0DQ44 A0A016TA97 Q3TLN7 A0A016U4Q3 A0A2P8YTE9 Q9XZG6 A0A1W4WVW2 A0A183TZ78 A0A0B2UVH4 M1EKM3 A0A2Y9DD68 A0A1U7R4S6 A0A140JWS1 A0A151N5U6 A0A1L8FCD4 T1JMA7 A0A1B0GN58 V9KKB5 E5SQL9 A0A3P4LVU6 A0A1L8F4P1 A0A016U4B9 M7BKI1 T1ISC0 F7IMS5 A0A2K6TFQ4 A0A0V1BC16 F7I0J5 A0A2K6G200 A0A0V0SHU4 A0A0V1BCN8 A0A1A8E027 A0A2R8NBR1 A0A1I8B3L8 A0A2P8XJ35 A0A091HTU2 A0A3B1JDI7 A0A2K6G203 A0A2K6FDD4 F7FL00 G1LSI9 A0A016TAD1 G3S576 Q3UUZ9 Q8VHB5 A0A3M0KBQ2 A0A3Q0D3V9 A0A3B4FV88 A0A087ZQS3 B3M1T1 A0A2Y9J8S9 A0A2K6RHV1 A0A1U7R3S6 A0A2I3G6J0 A0A3B1J929 A0A1A8PNH9 A0A2Y9E302 A0A2P2HXR5 A0A1B6GUM7 M3Y1M1 A2IBE1 A0A2U3VD21 A0A0A9X516 A0A1U7U5Z9 A0A2Y9JCI1 A0A146KNY1 A0A1D2NK15 A0A2U3YVE9 I3JGP7 A0A2R9C852 A0A2I3T6U9 A0A2J8T0E9 A0A2K6LTN4 A0A1A8RH30 L5KEW3 A0A1U7RKV7 G3SEJ4 G1SYS8 A0A3Q0GC63 A0A091GQF2
A0A0L7L7H9 A0A2W1C1Z7 A0A2A4IRL2 A0A2H1V6M7 A0A212F719 G3TMC4 V4AH85 A0A0R3PNH9 V3ZXK9 A0A1I8CXN0 H9JVL5 A0A0L8I5X5 A0A0K0DQ44 A0A016TA97 Q3TLN7 A0A016U4Q3 A0A2P8YTE9 Q9XZG6 A0A1W4WVW2 A0A183TZ78 A0A0B2UVH4 M1EKM3 A0A2Y9DD68 A0A1U7R4S6 A0A140JWS1 A0A151N5U6 A0A1L8FCD4 T1JMA7 A0A1B0GN58 V9KKB5 E5SQL9 A0A3P4LVU6 A0A1L8F4P1 A0A016U4B9 M7BKI1 T1ISC0 F7IMS5 A0A2K6TFQ4 A0A0V1BC16 F7I0J5 A0A2K6G200 A0A0V0SHU4 A0A0V1BCN8 A0A1A8E027 A0A2R8NBR1 A0A1I8B3L8 A0A2P8XJ35 A0A091HTU2 A0A3B1JDI7 A0A2K6G203 A0A2K6FDD4 F7FL00 G1LSI9 A0A016TAD1 G3S576 Q3UUZ9 Q8VHB5 A0A3M0KBQ2 A0A3Q0D3V9 A0A3B4FV88 A0A087ZQS3 B3M1T1 A0A2Y9J8S9 A0A2K6RHV1 A0A1U7R3S6 A0A2I3G6J0 A0A3B1J929 A0A1A8PNH9 A0A2Y9E302 A0A2P2HXR5 A0A1B6GUM7 M3Y1M1 A2IBE1 A0A2U3VD21 A0A0A9X516 A0A1U7U5Z9 A0A2Y9JCI1 A0A146KNY1 A0A1D2NK15 A0A2U3YVE9 I3JGP7 A0A2R9C852 A0A2I3T6U9 A0A2J8T0E9 A0A2K6LTN4 A0A1A8RH30 L5KEW3 A0A1U7RKV7 G3SEJ4 G1SYS8 A0A3Q0GC63 A0A091GQF2
PDB
5DVX
E-value=6.45804e-19,
Score=224
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cell membrane
Cell projection
Microvillus membrane
Cell projection
Microvillus membrane
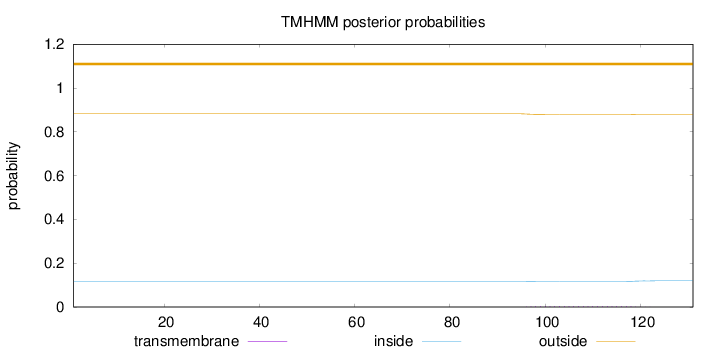
Length:
131
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09557
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.11787
outside
1 - 131
Population Genetic Test Statistics
Pi
209.009276
Theta
149.432322
Tajima's D
1.044498
CLR
0
CSRT
0.674116294185291
Interpretation
Uncertain
