Gene
KWMTBOMO03210
Pre Gene Modal
BGIBMGA013578
Annotation
PREDICTED:_carbonic_anhydrase_2-like_[Bombyx_mori]
Full name
Putative carbonic anhydrase 5
+ More
Carbonic anhydrase
Carbonic anhydrase
Alternative Name
Carbonate dehydratase 5
Carbonate dehydratase
Carbonate dehydratase
Location in the cell
Nuclear Reliability : 1.142
Sequence
CDS
ATGACTCGTGACGCCATTTTCGACAAGCACGAAGCCCATATACGCGGACCGCTGGTGATGAGAGGATACGGAGATGTTAAGGTCACAGCGATCAACAACGGACATACACTAAGATGGAAGGTGAAACCTGACACTGAGGCGCCAGTGCTCTCCGGGGGTCCACTTAGAGGGAACTACACCTTCCTGCAGTTTCATCTACACTGGCTCTCGGAACACGCTATTGACGGCATGAAATATCCCATGGAGATACACATGGTGCACGTGAAAACCGGTGTCCCGGTGGATGAAGCCTTGAAACGTCCAGATGGACTCGCCGTCATCGGCGTCATGTGTCAGATATACAGTAATGAAGAAAGCCAATTCGCTTTGGAGCAAATAATGCCCGCTGTACCGGACCTCGCAAGCAGATCTGAAGAAGAAACCGACCCTAAGAACATTGATTTAAGACGCCTGCTGAGTCCAAACCCTCAATCCTACTACACATACCACGGGTCCCTCACCACACCGGACTGCCAGGAATCAGTCACCTGGATCGTTATGGACAAACCTCTCATCATTGCTGACACACAGTATCAAGTCTTCAGCAAAGTCGACGTCGGTGGTCGAGACAACTATAGAAACTTGCAACAAGTCAACCGCGTCGTGTATCGCTCAGTAGCTTCTTGCTCCACGATCCTGATGCCGAGTTTCTTTGGAACCATAATTAGCTTGATCAGCTGCCTCTCGTCCTCCCTCACTGCTGGAGTCAACAAGGGATACTGCTACATCGTTAACATCAAGAGAAGATTGTTGGGCAGTACGGTGAAAAAGTGCGCGAAATTGGCGTAA
Protein
MTRDAIFDKHEAHIRGPLVMRGYGDVKVTAINNGHTLRWKVKPDTEAPVLSGGPLRGNYTFLQFHLHWLSEHAIDGMKYPMEIHMVHVKTGVPVDEALKRPDGLAVIGVMCQIYSNEESQFALEQIMPAVPDLASRSEEETDPKNIDLRRLLSPNPQSYYTYHGSLTTPDCQESVTWIVMDKPLIIADTQYQVFSKVDVGGRDNYRNLQQVNRVVYRSVASCSTILMPSFFGTIISLISCLSSSLTAGVNKGYCYIVNIKRRLLGSTVKKCAKLA
Summary
Catalytic Activity
H(+) + hydrogencarbonate = CO2 + H2O
Cofactor
Zn(2+)
Similarity
Belongs to the alpha-carbonic anhydrase family.
Keywords
Complete proteome
Lyase
Metal-binding
Reference proteome
Secreted
Signal
Zinc
Direct protein sequencing
Feature
chain Putative carbonic anhydrase 5
Uniprot
H9JVL5
A0A2A4IRL2
A0A2H1V6M7
A0A212F719
A0A0N1IHJ4
A0A194Q7X0
+ More
A0A2W1C1Z7 A0A2A4ISD7 A0A212F2G6 A0A2W1BSS8 A0A2I4C746 A0A146NR43 A0A0K0DQ44 E9FX48 B5X3I8 Q10462 A3FFY2 A0A060XWT2 B0K080 H3FHA2 A0A087XAG8 A0A3B3TI42 A0A3B3YRU7 A9QW25 A0A261CT67 G0N566 A0A3P8NSL8 A0A3P9CDM9 A0A261BDK7 A0A1A8HJQ0 E3LD70 A0A3P8TZ51 A0A3B4YTZ8 A0A1S3R2C2 I3JGP7 Q7T2K6 A0A2H2I757 R7UU94 K4NRP5 A0A3P8X094 I3JGP8 A0A2G5SVA9 A0A1A8DB90 A0A1A7ZF11 A0A1A8SD38 A0A1A8JBT8 A0A1A8PGI8 A0A0R3PNH9 A0A2G5SVS2 A0A3P8DER1 A0A3P9A0E3 A0A1B6J698 A0A3B1K2I1 A0A315W5C6 A0A091UNN1 A0A1I7TB76 A0A016TAD1 A0A3B4FV88 A0A3P8Y9W2 G3PPK1 A0A016TA97 A0A3B3YJM0 G3PPK5 A0A3B5Q5V9 A0A3B3QWB1 A0A0P5UU67 A0A2P8YTE9 A0A1W4WZE0 A0A3B5QPU6 A0A3B5LAD6 A0A162C6T7 A0A3B4THM8 A0A096M3S8 A0A087YKM6 A0A2U9BLY7 A0A0N8D3K6 A0A1B6FQP7 A0A3B1JLL4 A0A1A7Y143 A0A0P5AJF8 A0A2K5JT15 A0A2K5JT21 A0A0S7F752 Q92051 A0A088FII5 A0A3P9NPT1 A0A3B4YEI7 A0A0P5WCL3 A0A3B4EJX9
A0A2W1C1Z7 A0A2A4ISD7 A0A212F2G6 A0A2W1BSS8 A0A2I4C746 A0A146NR43 A0A0K0DQ44 E9FX48 B5X3I8 Q10462 A3FFY2 A0A060XWT2 B0K080 H3FHA2 A0A087XAG8 A0A3B3TI42 A0A3B3YRU7 A9QW25 A0A261CT67 G0N566 A0A3P8NSL8 A0A3P9CDM9 A0A261BDK7 A0A1A8HJQ0 E3LD70 A0A3P8TZ51 A0A3B4YTZ8 A0A1S3R2C2 I3JGP7 Q7T2K6 A0A2H2I757 R7UU94 K4NRP5 A0A3P8X094 I3JGP8 A0A2G5SVA9 A0A1A8DB90 A0A1A7ZF11 A0A1A8SD38 A0A1A8JBT8 A0A1A8PGI8 A0A0R3PNH9 A0A2G5SVS2 A0A3P8DER1 A0A3P9A0E3 A0A1B6J698 A0A3B1K2I1 A0A315W5C6 A0A091UNN1 A0A1I7TB76 A0A016TAD1 A0A3B4FV88 A0A3P8Y9W2 G3PPK1 A0A016TA97 A0A3B3YJM0 G3PPK5 A0A3B5Q5V9 A0A3B3QWB1 A0A0P5UU67 A0A2P8YTE9 A0A1W4WZE0 A0A3B5QPU6 A0A3B5LAD6 A0A162C6T7 A0A3B4THM8 A0A096M3S8 A0A087YKM6 A0A2U9BLY7 A0A0N8D3K6 A0A1B6FQP7 A0A3B1JLL4 A0A1A7Y143 A0A0P5AJF8 A0A2K5JT15 A0A2K5JT21 A0A0S7F752 Q92051 A0A088FII5 A0A3P9NPT1 A0A3B4YEI7 A0A0P5WCL3 A0A3B4EJX9
EC Number
4.2.1.1
Pubmed
EMBL
BABH01032447
NWSH01008790
PCG62415.1
ODYU01000955
SOQ36493.1
AGBW02009941
+ More
OWR49535.1 KQ460542 KPJ14143.1 KQ459579 KPI99505.1 KZ149911 PZC78133.1 PCG62416.1 AGBW02010745 OWR47922.1 PZC78132.1 GCES01152051 JAQ34271.1 GL732526 EFX88009.1 BT045607 BT058764 ACI33869.1 ACN10477.1 FO080190 EF375491 ABN51214.1 FR905752 CDQ81390.1 HE601482 CAP30217.4 AYCK01025318 EU273944 ABX71209.1 NIPN01000001 OZG25245.1 GL379839 EGT52986.1 NMWX01000001 OZG07736.1 HAEB01002559 HAEC01014668 SBQ82885.1 DS268407 LFJK02000005 EFO82916.1 POM51962.1 AERX01042727 AY307082 AB117757 FR904514 AAP73748.1 BAD36836.2 CDQ65658.1 AMQN01006271 KB297987 ELU09733.1 JX524150 AFV46145.1 PDUG01000006 PIC18960.1 HADZ01017639 HAEA01002168 SBQ30648.1 HADY01002583 HAEJ01009351 SBP41068.1 HAEH01002099 HAEI01013777 SBS16246.1 HAED01020652 HAEE01000852 SBR07273.1 HAEF01020435 HAEG01007777 SBR80139.1 UYYA01003961 VDM58193.1 PIC18961.1 UZAH01027748 VDO94652.1 GECU01013016 JAS94690.1 NHOQ01000342 PWA30799.1 KL409726 KFQ91520.1 JARK01001457 EYB99596.1 EYB99595.1 GDIP01125357 JAL78357.1 PYGN01000371 PSN47515.1 LRGB01001361 KZS12232.1 AYCK01001955 AYCK01001956 CP026249 AWP04678.1 AWP04679.1 AWP04680.1 AWP04681.1 AWP04682.1 AWP04683.1 AWP04684.1 AWP04685.1 AWP04686.1 GDIP01072199 JAM31516.1 GECZ01017308 JAS52461.1 HADW01011067 HADX01001483 SBP23715.1 GDIP01201244 JAJ22158.1 GBYX01470700 JAO10942.1 U55177 BC065611 KF650061 AIM43573.1 GDIP01087960 JAM15755.1
OWR49535.1 KQ460542 KPJ14143.1 KQ459579 KPI99505.1 KZ149911 PZC78133.1 PCG62416.1 AGBW02010745 OWR47922.1 PZC78132.1 GCES01152051 JAQ34271.1 GL732526 EFX88009.1 BT045607 BT058764 ACI33869.1 ACN10477.1 FO080190 EF375491 ABN51214.1 FR905752 CDQ81390.1 HE601482 CAP30217.4 AYCK01025318 EU273944 ABX71209.1 NIPN01000001 OZG25245.1 GL379839 EGT52986.1 NMWX01000001 OZG07736.1 HAEB01002559 HAEC01014668 SBQ82885.1 DS268407 LFJK02000005 EFO82916.1 POM51962.1 AERX01042727 AY307082 AB117757 FR904514 AAP73748.1 BAD36836.2 CDQ65658.1 AMQN01006271 KB297987 ELU09733.1 JX524150 AFV46145.1 PDUG01000006 PIC18960.1 HADZ01017639 HAEA01002168 SBQ30648.1 HADY01002583 HAEJ01009351 SBP41068.1 HAEH01002099 HAEI01013777 SBS16246.1 HAED01020652 HAEE01000852 SBR07273.1 HAEF01020435 HAEG01007777 SBR80139.1 UYYA01003961 VDM58193.1 PIC18961.1 UZAH01027748 VDO94652.1 GECU01013016 JAS94690.1 NHOQ01000342 PWA30799.1 KL409726 KFQ91520.1 JARK01001457 EYB99596.1 EYB99595.1 GDIP01125357 JAL78357.1 PYGN01000371 PSN47515.1 LRGB01001361 KZS12232.1 AYCK01001955 AYCK01001956 CP026249 AWP04678.1 AWP04679.1 AWP04680.1 AWP04681.1 AWP04682.1 AWP04683.1 AWP04684.1 AWP04685.1 AWP04686.1 GDIP01072199 JAM31516.1 GECZ01017308 JAS52461.1 HADW01011067 HADX01001483 SBP23715.1 GDIP01201244 JAJ22158.1 GBYX01470700 JAO10942.1 U55177 BC065611 KF650061 AIM43573.1 GDIP01087960 JAM15755.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000192220
+ More
UP000035642 UP000000305 UP000087266 UP000001940 UP000193380 UP000008549 UP000005239 UP000028760 UP000261500 UP000261480 UP000216463 UP000008068 UP000265100 UP000265160 UP000216624 UP000008281 UP000237256 UP000265080 UP000261400 UP000005207 UP000005237 UP000014760 UP000265120 UP000230233 UP000050601 UP000267027 UP000265140 UP000018467 UP000053283 UP000095282 UP000024635 UP000261460 UP000007635 UP000002852 UP000261540 UP000245037 UP000192223 UP000261380 UP000076858 UP000261420 UP000246464 UP000233080 UP000000437 UP000242638 UP000261360 UP000261440
UP000035642 UP000000305 UP000087266 UP000001940 UP000193380 UP000008549 UP000005239 UP000028760 UP000261500 UP000261480 UP000216463 UP000008068 UP000265100 UP000265160 UP000216624 UP000008281 UP000237256 UP000265080 UP000261400 UP000005207 UP000005237 UP000014760 UP000265120 UP000230233 UP000050601 UP000267027 UP000265140 UP000018467 UP000053283 UP000095282 UP000024635 UP000261460 UP000007635 UP000002852 UP000261540 UP000245037 UP000192223 UP000261380 UP000076858 UP000261420 UP000246464 UP000233080 UP000000437 UP000242638 UP000261360 UP000261440
Pfam
PF00194 Carb_anhydrase
Interpro
SUPFAM
SSF51069
SSF51069
Gene 3D
ProteinModelPortal
H9JVL5
A0A2A4IRL2
A0A2H1V6M7
A0A212F719
A0A0N1IHJ4
A0A194Q7X0
+ More
A0A2W1C1Z7 A0A2A4ISD7 A0A212F2G6 A0A2W1BSS8 A0A2I4C746 A0A146NR43 A0A0K0DQ44 E9FX48 B5X3I8 Q10462 A3FFY2 A0A060XWT2 B0K080 H3FHA2 A0A087XAG8 A0A3B3TI42 A0A3B3YRU7 A9QW25 A0A261CT67 G0N566 A0A3P8NSL8 A0A3P9CDM9 A0A261BDK7 A0A1A8HJQ0 E3LD70 A0A3P8TZ51 A0A3B4YTZ8 A0A1S3R2C2 I3JGP7 Q7T2K6 A0A2H2I757 R7UU94 K4NRP5 A0A3P8X094 I3JGP8 A0A2G5SVA9 A0A1A8DB90 A0A1A7ZF11 A0A1A8SD38 A0A1A8JBT8 A0A1A8PGI8 A0A0R3PNH9 A0A2G5SVS2 A0A3P8DER1 A0A3P9A0E3 A0A1B6J698 A0A3B1K2I1 A0A315W5C6 A0A091UNN1 A0A1I7TB76 A0A016TAD1 A0A3B4FV88 A0A3P8Y9W2 G3PPK1 A0A016TA97 A0A3B3YJM0 G3PPK5 A0A3B5Q5V9 A0A3B3QWB1 A0A0P5UU67 A0A2P8YTE9 A0A1W4WZE0 A0A3B5QPU6 A0A3B5LAD6 A0A162C6T7 A0A3B4THM8 A0A096M3S8 A0A087YKM6 A0A2U9BLY7 A0A0N8D3K6 A0A1B6FQP7 A0A3B1JLL4 A0A1A7Y143 A0A0P5AJF8 A0A2K5JT15 A0A2K5JT21 A0A0S7F752 Q92051 A0A088FII5 A0A3P9NPT1 A0A3B4YEI7 A0A0P5WCL3 A0A3B4EJX9
A0A2W1C1Z7 A0A2A4ISD7 A0A212F2G6 A0A2W1BSS8 A0A2I4C746 A0A146NR43 A0A0K0DQ44 E9FX48 B5X3I8 Q10462 A3FFY2 A0A060XWT2 B0K080 H3FHA2 A0A087XAG8 A0A3B3TI42 A0A3B3YRU7 A9QW25 A0A261CT67 G0N566 A0A3P8NSL8 A0A3P9CDM9 A0A261BDK7 A0A1A8HJQ0 E3LD70 A0A3P8TZ51 A0A3B4YTZ8 A0A1S3R2C2 I3JGP7 Q7T2K6 A0A2H2I757 R7UU94 K4NRP5 A0A3P8X094 I3JGP8 A0A2G5SVA9 A0A1A8DB90 A0A1A7ZF11 A0A1A8SD38 A0A1A8JBT8 A0A1A8PGI8 A0A0R3PNH9 A0A2G5SVS2 A0A3P8DER1 A0A3P9A0E3 A0A1B6J698 A0A3B1K2I1 A0A315W5C6 A0A091UNN1 A0A1I7TB76 A0A016TAD1 A0A3B4FV88 A0A3P8Y9W2 G3PPK1 A0A016TA97 A0A3B3YJM0 G3PPK5 A0A3B5Q5V9 A0A3B3QWB1 A0A0P5UU67 A0A2P8YTE9 A0A1W4WZE0 A0A3B5QPU6 A0A3B5LAD6 A0A162C6T7 A0A3B4THM8 A0A096M3S8 A0A087YKM6 A0A2U9BLY7 A0A0N8D3K6 A0A1B6FQP7 A0A3B1JLL4 A0A1A7Y143 A0A0P5AJF8 A0A2K5JT15 A0A2K5JT21 A0A0S7F752 Q92051 A0A088FII5 A0A3P9NPT1 A0A3B4YEI7 A0A0P5WCL3 A0A3B4EJX9
PDB
5G0B
E-value=1.35507e-28,
Score=313
Ontologies
GO
PANTHER
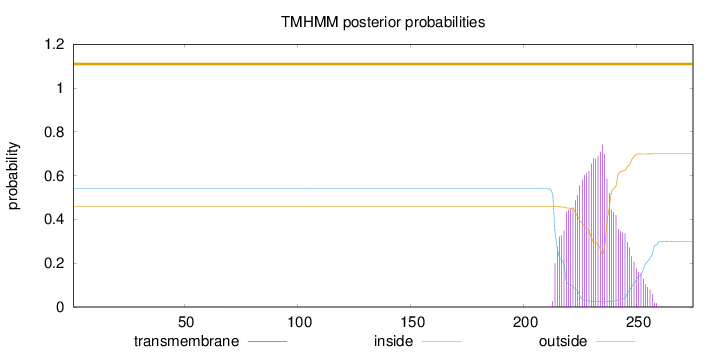
Topology
Subcellular location
Secreted
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.89387
Exp number, first 60 AAs:
0.00188
Total prob of N-in:
0.54033
outside
1 - 275
Population Genetic Test Statistics
Pi
222.238595
Theta
150.648547
Tajima's D
1.120958
CLR
0.480949
CSRT
0.700064996750163
Interpretation
Uncertain
