Gene
KWMTBOMO03208
Pre Gene Modal
BGIBMGA013576
Annotation
PREDICTED:_organic_cation/carnitine_transporter_7-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.927
Sequence
CDS
ATGCTCTGCAGTAGCATCATAGCAGCCATGGTCTTCGAGCTTTACTCCGTATCATATCTCGTGCCGGCTAGTGCCTGCGAGCTGAACACCACCAGTACGCAGCAAGGATTGATGGCTGGTCTCCCATTAATCGGTGTGATCACAACCTCACACATTTGGGGGTATTTAGCCGACACTCGAGGCAGAAAGAAGATGCTCTGCATTTCCATGATCATTGGCTTCGTGGCTGGTTCGTCAGCAGCCCTGGCCCCCAACTGGATCGTGCTTAGCATACTAAAGTTCCTGTCTTCGGCTGCGGTATCGGGATCATCAGCGTTGGCTTTAGCACTGATAGGGGAGTGCACTCCTGCGATACGCCGGAGTACCATGCTGCTACTCACCACCAGCGCGCTGCTATCGAGCACCGGGGTCATGGCCATACTCTCGATACCGATCCTGCCAATGAAGTTCTCCTACTACATACCGATCCTGAACATCCACTTCAACTCCTGGCGCCTTCTAAATGTGATCTTCAGTCTGCCGTGTGCTCTAAACGCGATCGGTTCGATGTGCTGTTATGAAAGTCCAAAGTATTTGCTGAGCGTTGGAGAAGATGAAAAAGCGTTGGAGATCCTTCAGGGAATTTTTAAGCATAACAGCGGAAAGAGCGCAGATTTATACCAGGTGGAGTCAATAGTTCTAGATGAAGATAACGCGAATAAGAAAGCCAAGGGTTTCTGGGAGGTGCTTGCCGAGCAGACGCTACCTTTATTCAAACCACCGCTACTGAAGAGTACGCTATTACTCGGTTTCATGTTCATCTTAGTTTTCATTTGCATAAACTCATTCTTAGTTTGGCTTCCATTCATGGTGGACGCTTTTATGTCTTCGGTGGAGAAAGGATACTCCGGCATGACCATGTGCGAAATGATTAGGGTGGCCAGGAATGAAACCGACCTGGAAGAGACAAATTCCACTTGCTCTATGAACAGAGTGGCCATGACCATGGTTTTCGGTATCTGCATAGTTCTGGCCGTCTTGAACGTGGTGGCTAGTGGCGTAATCAGCATCATAGGCAAGAGGAGACTATTTATAGGCATACAGGTGATGGCCGGTGTAAGCAGTATCTTACTGAACGTGAGCTCCATGTGGGTAGTAAGCGCTCTTCTCTTCATAGCGGTTCTATCTGATGTCATCAACTTTGGATTCCTGTCAGCATTCGCTGTGGACCTGTATCCAACTTTTGTGAAGGCGAAGGCCATATGCCTGATGGTGATGCTGGGTCGCGGCAGCACGATCCTCGGCATCAACGTGCTCAAGAGCCTCATCGAGACGAACTGCGAAGCTGCCTTTTATCTATTCGGAGGGATCACTCTAGCCGGTGGACTCTTCGGATTCCTGCTTCCTCCAGACGGGAATCTATTCAAACAGAAAACTAAAATCTAA
Protein
MLCSSIIAAMVFELYSVSYLVPASACELNTTSTQQGLMAGLPLIGVITTSHIWGYLADTRGRKKMLCISMIIGFVAGSSAALAPNWIVLSILKFLSSAAVSGSSALALALIGECTPAIRRSTMLLLTTSALLSSTGVMAILSIPILPMKFSYYIPILNIHFNSWRLLNVIFSLPCALNAIGSMCCYESPKYLLSVGEDEKALEILQGIFKHNSGKSADLYQVESIVLDEDNANKKAKGFWEVLAEQTLPLFKPPLLKSTLLLGFMFILVFICINSFLVWLPFMVDAFMSSVEKGYSGMTMCEMIRVARNETDLEETNSTCSMNRVAMTMVFGICIVLAVLNVVASGVISIIGKRRLFIGIQVMAGVSSILLNVSSMWVVSALLFIAVLSDVINFGFLSAFAVDLYPTFVKAKAICLMVMLGRGSTILGINVLKSLIETNCEAAFYLFGGITLAGGLFGFLLPPDGNLFKQKTKI
Summary
Uniprot
H9JVL3
A0A2H1V0R1
A0A2H1VUK7
A0A2A4JT62
A0A2W1BZI9
A0A194Q2E7
+ More
A0A212FI93 A0A2W1BWW5 A0A0N1I9N3 A0A194Q2D2 A0A0N1IG89 A0A2H1WI98 A0A2A4JMQ7 A0A212F5P7 A0A2A4JLI7 A0A2A4JLT5 A0A2H1WGI5 A0A2H1V250 A0A194Q3A9 A0A212EY43 A0A2W1BUU8 A0A212EY03 A0A0N1I9M8 A0A0N0PCN2 A0A194QN61 A0A2A4K5Q7 A0A194PP30 A0A2A4JLN1 A0A194Q1T6 A0A2A4JKW1 A0A0N1IPG1 A0A2H1W801 A0A2H1W7K7 A0A194Q1R5 H9JVM2 A0A212EYD8 H9JVR6 A0A194PJI3 A0A194Q1T1 A0A2A4JX40 A0A212EQC0 A0A194QN48 H9JVR7 A0A2H1VUC9 A0A2H1WYY1 A0A2H1X0I9 A0A194Q1Q2 A0A194Q7W0 A0A212EZN8 A0A0L7KD47 H9JV72 A0A2H1WYT6 A0A2A4J999 A0A194Q800 A0A212F5C2 W8AY09 A0A034VLH4 A0A1J1HSM7 A0A0K8UH50 A0A034VFR0 U5ETH9 W8C469 A0A0K8U925 A0A336KU37 H9JVL1 A0A0N0PCK2 A0A0A1XKB5 B0WKE4 A0A0A1XE46 A0A2A4JAH3 A0A1L8EFQ6 A0A1L8EFU2 A0A1L8EFY3 A0A2H1WJU4 A0A1Q3FLR3 Q6U1H2 A0A1I8P1U8 A0A1I8MVX6 A0A1I8PH06 A0A1I8MVY3 A0A1J1HFZ0 A0A1I8P1R5 T1PCW5 T1P8E1 A0A182YEX8 A0A1B6G7N2 A0A2W1BGE8 B0W415 A0A1Q3FR20 A0A0L0BWY3 A0A1Q3FRJ7 A0A2H1WNU1 A0A1B6I2U3
A0A212FI93 A0A2W1BWW5 A0A0N1I9N3 A0A194Q2D2 A0A0N1IG89 A0A2H1WI98 A0A2A4JMQ7 A0A212F5P7 A0A2A4JLI7 A0A2A4JLT5 A0A2H1WGI5 A0A2H1V250 A0A194Q3A9 A0A212EY43 A0A2W1BUU8 A0A212EY03 A0A0N1I9M8 A0A0N0PCN2 A0A194QN61 A0A2A4K5Q7 A0A194PP30 A0A2A4JLN1 A0A194Q1T6 A0A2A4JKW1 A0A0N1IPG1 A0A2H1W801 A0A2H1W7K7 A0A194Q1R5 H9JVM2 A0A212EYD8 H9JVR6 A0A194PJI3 A0A194Q1T1 A0A2A4JX40 A0A212EQC0 A0A194QN48 H9JVR7 A0A2H1VUC9 A0A2H1WYY1 A0A2H1X0I9 A0A194Q1Q2 A0A194Q7W0 A0A212EZN8 A0A0L7KD47 H9JV72 A0A2H1WYT6 A0A2A4J999 A0A194Q800 A0A212F5C2 W8AY09 A0A034VLH4 A0A1J1HSM7 A0A0K8UH50 A0A034VFR0 U5ETH9 W8C469 A0A0K8U925 A0A336KU37 H9JVL1 A0A0N0PCK2 A0A0A1XKB5 B0WKE4 A0A0A1XE46 A0A2A4JAH3 A0A1L8EFQ6 A0A1L8EFU2 A0A1L8EFY3 A0A2H1WJU4 A0A1Q3FLR3 Q6U1H2 A0A1I8P1U8 A0A1I8MVX6 A0A1I8PH06 A0A1I8MVY3 A0A1J1HFZ0 A0A1I8P1R5 T1PCW5 T1P8E1 A0A182YEX8 A0A1B6G7N2 A0A2W1BGE8 B0W415 A0A1Q3FR20 A0A0L0BWY3 A0A1Q3FRJ7 A0A2H1WNU1 A0A1B6I2U3
Pubmed
EMBL
BABH01032453
ODYU01000151
SOQ34435.1
ODYU01004537
SOQ44529.1
NWSH01000659
+ More
PCG74966.1 KZ149911 PZC78136.1 KQ459579 KPI99508.1 AGBW02008417 OWR53448.1 PZC78135.1 KQ460542 KPJ14146.1 KPI99493.1 KPJ14129.1 ODYU01008516 SOQ52184.1 NWSH01001089 PCG72680.1 AGBW02010155 OWR49057.1 PCG72679.1 PCG72678.1 SOQ52183.1 SOQ34432.1 KPI99494.1 AGBW02011638 OWR46364.1 PZC78141.1 AGBW02011653 OWR46341.1 KPJ14130.1 KPJ14131.1 KQ461194 KPJ06947.1 NWSH01000098 PCG79585.1 KQ459603 KPI92885.1 PCG72686.1 KPI99511.1 PCG72687.1 KPJ14127.1 ODYU01006861 SOQ49086.1 SOQ49085.1 KPI99491.1 BABH01032396 BABH01032397 BABH01032398 BABH01032399 AGBW02011531 OWR46513.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 KPI92884.1 KPI99506.1 NWSH01000457 PCG76268.1 AGBW02013272 OWR43679.1 KPJ06948.1 BABH01032426 BABH01032427 BABH01032428 BABH01032429 ODYU01004504 SOQ44451.1 ODYU01012071 SOQ58176.1 ODYU01012422 SOQ58716.1 KPI99507.1 KPI99495.1 AGBW02011241 OWR46942.1 JTDY01010086 KOB61253.1 BABH01037389 ODYU01012108 SOQ58229.1 NWSH01002380 PCG68429.1 KPI99535.1 AGBW02010212 OWR48924.1 GAMC01012655 JAB93900.1 GAKP01015980 JAC42972.1 CVRI01000020 CRK91005.1 GDHF01026325 JAI25989.1 GAKP01018554 JAC40398.1 GANO01004365 JAB55506.1 GAMC01009766 JAB96789.1 GDHF01029221 JAI23093.1 UFQS01000919 UFQT01000919 SSX07668.1 SSX28006.1 BABH01032458 KPJ14144.1 GBXI01002836 JAD11456.1 DS231971 EDS29763.1 GBXI01004603 JAD09689.1 PCG68430.1 GFDG01001242 JAV17557.1 GFDG01001241 JAV17558.1 GFDG01001245 JAV17554.1 ODYU01009141 SOQ53353.1 GFDL01006485 JAV28560.1 AY380837 AAQ88394.1 CVRI01000002 CRK86895.1 KA646499 AFP61128.1 KA644899 AFP59528.1 GECZ01011345 JAS58424.1 KZ150202 PZC72277.1 DS231834 EDS32608.1 GFDL01005008 JAV30037.1 JRES01001303 KNC23749.1 GFDL01004992 JAV30053.1 ODYU01009931 SOQ54677.1 GECU01026455 JAS81251.1
PCG74966.1 KZ149911 PZC78136.1 KQ459579 KPI99508.1 AGBW02008417 OWR53448.1 PZC78135.1 KQ460542 KPJ14146.1 KPI99493.1 KPJ14129.1 ODYU01008516 SOQ52184.1 NWSH01001089 PCG72680.1 AGBW02010155 OWR49057.1 PCG72679.1 PCG72678.1 SOQ52183.1 SOQ34432.1 KPI99494.1 AGBW02011638 OWR46364.1 PZC78141.1 AGBW02011653 OWR46341.1 KPJ14130.1 KPJ14131.1 KQ461194 KPJ06947.1 NWSH01000098 PCG79585.1 KQ459603 KPI92885.1 PCG72686.1 KPI99511.1 PCG72687.1 KPJ14127.1 ODYU01006861 SOQ49086.1 SOQ49085.1 KPI99491.1 BABH01032396 BABH01032397 BABH01032398 BABH01032399 AGBW02011531 OWR46513.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 KPI92884.1 KPI99506.1 NWSH01000457 PCG76268.1 AGBW02013272 OWR43679.1 KPJ06948.1 BABH01032426 BABH01032427 BABH01032428 BABH01032429 ODYU01004504 SOQ44451.1 ODYU01012071 SOQ58176.1 ODYU01012422 SOQ58716.1 KPI99507.1 KPI99495.1 AGBW02011241 OWR46942.1 JTDY01010086 KOB61253.1 BABH01037389 ODYU01012108 SOQ58229.1 NWSH01002380 PCG68429.1 KPI99535.1 AGBW02010212 OWR48924.1 GAMC01012655 JAB93900.1 GAKP01015980 JAC42972.1 CVRI01000020 CRK91005.1 GDHF01026325 JAI25989.1 GAKP01018554 JAC40398.1 GANO01004365 JAB55506.1 GAMC01009766 JAB96789.1 GDHF01029221 JAI23093.1 UFQS01000919 UFQT01000919 SSX07668.1 SSX28006.1 BABH01032458 KPJ14144.1 GBXI01002836 JAD11456.1 DS231971 EDS29763.1 GBXI01004603 JAD09689.1 PCG68430.1 GFDG01001242 JAV17557.1 GFDG01001241 JAV17558.1 GFDG01001245 JAV17554.1 ODYU01009141 SOQ53353.1 GFDL01006485 JAV28560.1 AY380837 AAQ88394.1 CVRI01000002 CRK86895.1 KA646499 AFP61128.1 KA644899 AFP59528.1 GECZ01011345 JAS58424.1 KZ150202 PZC72277.1 DS231834 EDS32608.1 GFDL01005008 JAV30037.1 JRES01001303 KNC23749.1 GFDL01004992 JAV30053.1 ODYU01009931 SOQ54677.1 GECU01026455 JAS81251.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JVL3
A0A2H1V0R1
A0A2H1VUK7
A0A2A4JT62
A0A2W1BZI9
A0A194Q2E7
+ More
A0A212FI93 A0A2W1BWW5 A0A0N1I9N3 A0A194Q2D2 A0A0N1IG89 A0A2H1WI98 A0A2A4JMQ7 A0A212F5P7 A0A2A4JLI7 A0A2A4JLT5 A0A2H1WGI5 A0A2H1V250 A0A194Q3A9 A0A212EY43 A0A2W1BUU8 A0A212EY03 A0A0N1I9M8 A0A0N0PCN2 A0A194QN61 A0A2A4K5Q7 A0A194PP30 A0A2A4JLN1 A0A194Q1T6 A0A2A4JKW1 A0A0N1IPG1 A0A2H1W801 A0A2H1W7K7 A0A194Q1R5 H9JVM2 A0A212EYD8 H9JVR6 A0A194PJI3 A0A194Q1T1 A0A2A4JX40 A0A212EQC0 A0A194QN48 H9JVR7 A0A2H1VUC9 A0A2H1WYY1 A0A2H1X0I9 A0A194Q1Q2 A0A194Q7W0 A0A212EZN8 A0A0L7KD47 H9JV72 A0A2H1WYT6 A0A2A4J999 A0A194Q800 A0A212F5C2 W8AY09 A0A034VLH4 A0A1J1HSM7 A0A0K8UH50 A0A034VFR0 U5ETH9 W8C469 A0A0K8U925 A0A336KU37 H9JVL1 A0A0N0PCK2 A0A0A1XKB5 B0WKE4 A0A0A1XE46 A0A2A4JAH3 A0A1L8EFQ6 A0A1L8EFU2 A0A1L8EFY3 A0A2H1WJU4 A0A1Q3FLR3 Q6U1H2 A0A1I8P1U8 A0A1I8MVX6 A0A1I8PH06 A0A1I8MVY3 A0A1J1HFZ0 A0A1I8P1R5 T1PCW5 T1P8E1 A0A182YEX8 A0A1B6G7N2 A0A2W1BGE8 B0W415 A0A1Q3FR20 A0A0L0BWY3 A0A1Q3FRJ7 A0A2H1WNU1 A0A1B6I2U3
A0A212FI93 A0A2W1BWW5 A0A0N1I9N3 A0A194Q2D2 A0A0N1IG89 A0A2H1WI98 A0A2A4JMQ7 A0A212F5P7 A0A2A4JLI7 A0A2A4JLT5 A0A2H1WGI5 A0A2H1V250 A0A194Q3A9 A0A212EY43 A0A2W1BUU8 A0A212EY03 A0A0N1I9M8 A0A0N0PCN2 A0A194QN61 A0A2A4K5Q7 A0A194PP30 A0A2A4JLN1 A0A194Q1T6 A0A2A4JKW1 A0A0N1IPG1 A0A2H1W801 A0A2H1W7K7 A0A194Q1R5 H9JVM2 A0A212EYD8 H9JVR6 A0A194PJI3 A0A194Q1T1 A0A2A4JX40 A0A212EQC0 A0A194QN48 H9JVR7 A0A2H1VUC9 A0A2H1WYY1 A0A2H1X0I9 A0A194Q1Q2 A0A194Q7W0 A0A212EZN8 A0A0L7KD47 H9JV72 A0A2H1WYT6 A0A2A4J999 A0A194Q800 A0A212F5C2 W8AY09 A0A034VLH4 A0A1J1HSM7 A0A0K8UH50 A0A034VFR0 U5ETH9 W8C469 A0A0K8U925 A0A336KU37 H9JVL1 A0A0N0PCK2 A0A0A1XKB5 B0WKE4 A0A0A1XE46 A0A2A4JAH3 A0A1L8EFQ6 A0A1L8EFU2 A0A1L8EFY3 A0A2H1WJU4 A0A1Q3FLR3 Q6U1H2 A0A1I8P1U8 A0A1I8MVX6 A0A1I8PH06 A0A1I8MVY3 A0A1J1HFZ0 A0A1I8P1R5 T1PCW5 T1P8E1 A0A182YEX8 A0A1B6G7N2 A0A2W1BGE8 B0W415 A0A1Q3FR20 A0A0L0BWY3 A0A1Q3FRJ7 A0A2H1WNU1 A0A1B6I2U3
PDB
4LDS
E-value=0.0166492,
Score=90
Ontologies
GO
Topology
SignalP
Position: 1 - 25,
Likelihood: 0.506842
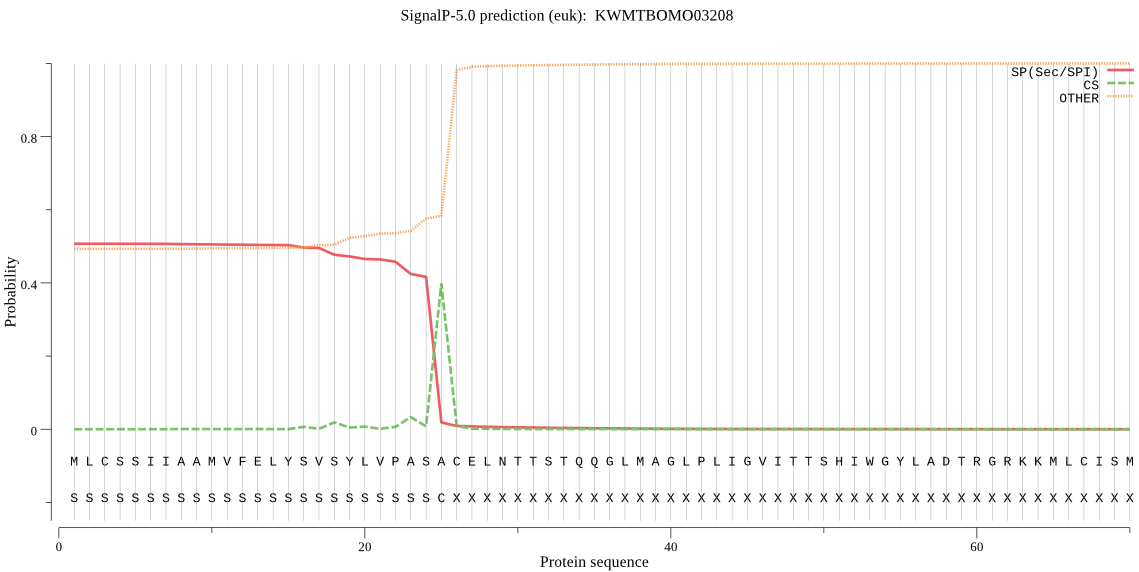
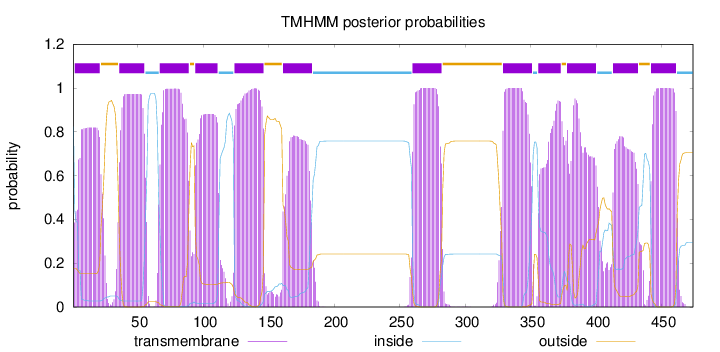
Length:
474
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
234.10883
Exp number, first 60 AAs:
37.15481
Total prob of N-in:
0.82441
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 21
outside
22 - 35
TMhelix
36 - 55
inside
56 - 66
TMhelix
67 - 89
outside
90 - 93
TMhelix
94 - 111
inside
112 - 123
TMhelix
124 - 146
outside
147 - 160
TMhelix
161 - 183
inside
184 - 259
TMhelix
260 - 282
outside
283 - 328
TMhelix
329 - 351
inside
352 - 355
TMhelix
356 - 373
outside
374 - 377
TMhelix
378 - 400
inside
401 - 412
TMhelix
413 - 432
outside
433 - 441
TMhelix
442 - 461
inside
462 - 474
Population Genetic Test Statistics
Pi
195.445126
Theta
150.807126
Tajima's D
0.92499
CLR
0.449275
CSRT
0.636118194090295
Interpretation
Uncertain
