Gene
KWMTBOMO03207
Pre Gene Modal
BGIBMGA013640
Annotation
PREDICTED:_chorion_peroxidase_[Bombyx_mori]
Full name
Chorion peroxidase
Alternative Name
Peroxinectin-related protein
Location in the cell
PlasmaMembrane Reliability : 1.525
Sequence
CDS
ATGAATTTCAAAAGCTTCATTTTGCTCTTAATTAATGTGTTCTTATTGAGTGTCATTGAGGATAGTTCTTGTCACGTTTACGAAGGCAGCGCACGTGAGGCCTCAGAACACATTCACGTACAAGAGCGACCATGCGCCGTCTGCCCCAGAGGCGGGAAGTGTGTCCCGAAGGTCAAATGTCCGGCCCACGTCAGACCGGGATCGTTGAATCCGACCTGCCACTTGGTCCTTGGACATATCGGTATCTGCTGCTTCACCGGGCAAAAGCATTCAGCCCAATTAGAATCTACTCAGCGTTCTGGCGCCATCAGTATAGAAGACATTAAAAGCAGCCATGACGTATCAAGGCAGAAAATGGTCCAGTGGATAGCGAACGAGAAAGTTCTAGAACGTTCTGCTGATACAATCGTCAGTAGTTCGGCACCTAGTTACGGGCATCATTTGTCCATGGTTACGTACGACAAGAGAGTTGAGGGTCTTGGACGTGGTGGACTGTTGAACGTCTTCGCTGCTCTGGAACTGAAGTCTCGGCAGGCGGTCTCTGATGATGAGCTGGAGTTGGGAGCAACAGGACATACTGATGGTCCCTTTTGCCCTAAGCCCCCGCAGTGCCCGGATACCCAAAGCCGGTACAGATCTATAGATGGGGAGTGCAACAATCTGGCCAACCCAACGTGGGGAGCTGTCAATACTGGCTTCGAGAGGCTTCTACCACCAGATTATAGTGACGGCGTGTGGGCAATGAGAGTCTCCGCAGCAGGTAATCCCCTTCCGAGCGCGAGGGTGGTCAGCAGCGTTCTCCTGCCAGAAGGAAACCATCCAAGTCCCACACACAACCTCATGTTCATGCAGTTTGGACAGTTTATAGCACACGACACCAGCGCTGGAGTTATGTTCGCATCGGGTAACAACACTGGAATATCCTGTTGCGCTGAAGACGGTGTGGACCAGCTGCATCCGAAGCAGCAGCACTGGGCCTGTGCTCCAATCACCGCAGTTCCCGATGACCCTTTCTATGGTTTCTTTGGTCAGAAATGCCTCAATTTCGTGCGCACACAACTCGCACCAGCCAGCGATTGTTCCGTTGGCTATGCCAAGCAAATGAATGGCGCCACGCATTACCCTGATTTGTCTCACTTATACGGCACCTTTCCCGAAAAACTCTCATTGGTGCGCGGCGAGGGAGGGTTTCTGAAGACATTCAATGACTTTGGCAGAGCCCTCCCACCTTTGACTAAGAGAAGAGAATGCGTTAATATGGATGGTGGTAGCCCTTGCTTTGAATCAGGAGACAGTCACGGGAATCAGTTCATCTCACTGACAGCATTCCACACGTTATGGTCGAGGGAGCACAACCGAGTCGCGCAAGCCCTGTCGAGGTTGAACCCTCATTGGGACGAAGACACAGTCTTTATGGAGACCAGAAGGATACTGCAGGCAGAATTCCAGCATATCATTTATAATGAGTGGCTTCCTCTATTGATAGGTCACCAAATGATGCAGCTCTTCAATTTATCTCCATCTTCAGAGTATTCCTCGTCGTACGATCCAACAGTCAATCCGTCTATTACAGCAGAATTCGCGACCGCAGCCATGAGATTTGGCCACTCCATTGTTGATGGAAGAATTGTAATTCCCAACACGAAAACAGGAGAAGTTCACGAGACCATATCGATACCGGAAGTGATGTTTCAGCCGTCGAGAATGCGGCTACGTCACTTCTTGGATCGGCTGCTGATTGGCCTGACGCTGCAACCGATGCAGAGTGTCGATCCGTTTATCTCTGAAGGGCTTACGTCCTACATGTTCCGTGGCACCAACCCCTATGGCCTGGATCTAGCGTCAATAAACATTCAACGCGGCAGAGACTACGGAGTCAGATCCTACAATAGCTACCGGCGCTTGTGTGGGCTTCAGCCATTTGAAAGCTTCGAGCAATTTCCACAAAGTGCTGCAAAGCGTCTAGCGTCTGTCTACGAGAGTCCGGAAGACATAGACCTTTGGGTTGGAGGTCTGCTGGAGGCGCCGATGGAAGAGGCAGTTATCGGGCCAACTTTCGCTCATATCATAGCCGACCAATTCTACAGGCTCAAGGCTGGAGATAGATACTTCTACGATAATGGTCCTGATATAAATCCCGGCGCTTTTACTCCAAGCCAACTGACAGAAATAAAGAAGGTTAAACTATCAAGACTGATTTGTGACAATAGCGATGGTATAGAACTTGTAACCCAGCCAGTTGAAGCCTTTTATAGAGCAGACCTACCAGGAAATGAATTAGTGGCTTGCAACAGTGGACGCATACCTTTTATGGATTTGAGCAGATTTAAGGCGCTCAAAACTTAA
Protein
MNFKSFILLLINVFLLSVIEDSSCHVYEGSAREASEHIHVQERPCAVCPRGGKCVPKVKCPAHVRPGSLNPTCHLVLGHIGICCFTGQKHSAQLESTQRSGAISIEDIKSSHDVSRQKMVQWIANEKVLERSADTIVSSSAPSYGHHLSMVTYDKRVEGLGRGGLLNVFAALELKSRQAVSDDELELGATGHTDGPFCPKPPQCPDTQSRYRSIDGECNNLANPTWGAVNTGFERLLPPDYSDGVWAMRVSAAGNPLPSARVVSSVLLPEGNHPSPTHNLMFMQFGQFIAHDTSAGVMFASGNNTGISCCAEDGVDQLHPKQQHWACAPITAVPDDPFYGFFGQKCLNFVRTQLAPASDCSVGYAKQMNGATHYPDLSHLYGTFPEKLSLVRGEGGFLKTFNDFGRALPPLTKRRECVNMDGGSPCFESGDSHGNQFISLTAFHTLWSREHNRVAQALSRLNPHWDEDTVFMETRRILQAEFQHIIYNEWLPLLIGHQMMQLFNLSPSSEYSSSYDPTVNPSITAEFATAAMRFGHSIVDGRIVIPNTKTGEVHETISIPEVMFQPSRMRLRHFLDRLLIGLTLQPMQSVDPFISEGLTSYMFRGTNPYGLDLASINIQRGRDYGVRSYNSYRRLCGLQPFESFEQFPQSAAKRLASVYESPEDIDLWVGGLLEAPMEEAVIGPTFAHIIADQFYRLKAGDRYFYDNGPDINPGAFTPSQLTEIKKVKLSRLICDNSDGIELVTQPVEAFYRADLPGNELVACNSGRIPFMDLSRFKALKT
Summary
Description
Required for ovarian follicle maturation. Involved in the formation of a rigid and insoluble egg chorion by catalyzing chorion protein cross-linking through dityrosine formation and phenol oxidase-catalyzed chorion melanization.
Involved in the formation of a rigid and insoluble egg chorion by catalyzing chorion protein cross-linking through dityrosine formation and phenol oxidase-catalyzed chorion melanization.
Involved in the formation of a rigid and insoluble egg chorion by catalyzing chorion protein cross-linking through dityrosine formation and phenol oxidase-catalyzed chorion melanization.
Catalytic Activity
2 a phenolic donor + H2O2 = 2 a phenolic radical donor + 2 H2O
Cofactor
heme b
Subunit
Heterodimer.
Similarity
Belongs to the peroxidase family. XPO subfamily.
Keywords
Acetylation
Complete proteome
Disulfide bond
Glycoprotein
Heme
Hydrogen peroxide
Iron
Metal-binding
Oxidoreductase
Peroxidase
Reference proteome
Secreted
Signal
Cleavage on pair of basic residues
Hydroxylation
Feature
chain Chorion peroxidase
Uniprot
A0A348B065
H9JVS7
A0A2H1WFB0
A0A2H1V2D9
A0A2W1BY28
A0A2A4JSF2
+ More
A0A194Q3C2 A0A0N0P9N7 A0A0N1PI15 A0A139WMA7 A0A1Y1MHD3 A0A1W4V5J1 B3P0H0 B4PKH9 B4QV50 Q9VEG6 B4IBE0 U4UDX4 N6TJI6 B5DM11 A0A3B0KPN2 A0A0K8VB63 B4NFK6 A0A034VZV8 B3MTN9 A0A1B0CFZ9 A0A1J1HL34 A0A0L0BQN4 A0A1I8PRJ4 W8CED3 A0A0A1WGZ5 A0A1L8E461 A0A034W3E1 A0A1L8E467 A0A1I8MYK6 A0A0M4F596 U5ES00 A0A336KIB1 B4M5G6 B4JUF8 B4KDQ0 A0A1B0DRI5 A0A182JWM4 A0A182QE50 T1DNT2 A0A182M8J6 W5JFF7 A0A182RSK4 A0A182VX07 A0A2P2IA20 A0A182I859 Q7QH73 A0A182UH95 A0A182P440 A0A182V5S6 A0A182S8Q9 A0A182YF53 A0A182XFE3 A0A182L7T6 A0A182J1F9 A0A182NG33 A0A084VTJ6 A0A182YF54 A0A1Q3EV39 A0A0N8DGW9 A0A0N8EED2 A0A0P5Y961 A0A162Q098 A0A0P5I633 A0A182GM26 B0WKK0 E9GJ60 A0A0P4ZMN0 A0A0P5G0R9 A0A182MTG7 A0A0P6DHJ3 A0A0P5ULU2 A0A0P5SG58 A0A0P5RV48 A0A0N8EE02 A0A0P5LZK4 A0A0P5GUL0 A0A0P5BQS1 Q17CY5 A0A1S4F7F6 A0A0P4ZXK5 A0A0P5S3L9 A0A0P5M7P6 A0A0P5IEW1 A0A0N7ZV01 A0A0P5TPL4 A0A162DAW2 A0A0P5EVV2 A0A0P5AI81 A0A182P439 A0A0P6JNR6 A0A0N7ZU83 A0A0P5BVJ2 A0A0P6EWY1
A0A194Q3C2 A0A0N0P9N7 A0A0N1PI15 A0A139WMA7 A0A1Y1MHD3 A0A1W4V5J1 B3P0H0 B4PKH9 B4QV50 Q9VEG6 B4IBE0 U4UDX4 N6TJI6 B5DM11 A0A3B0KPN2 A0A0K8VB63 B4NFK6 A0A034VZV8 B3MTN9 A0A1B0CFZ9 A0A1J1HL34 A0A0L0BQN4 A0A1I8PRJ4 W8CED3 A0A0A1WGZ5 A0A1L8E461 A0A034W3E1 A0A1L8E467 A0A1I8MYK6 A0A0M4F596 U5ES00 A0A336KIB1 B4M5G6 B4JUF8 B4KDQ0 A0A1B0DRI5 A0A182JWM4 A0A182QE50 T1DNT2 A0A182M8J6 W5JFF7 A0A182RSK4 A0A182VX07 A0A2P2IA20 A0A182I859 Q7QH73 A0A182UH95 A0A182P440 A0A182V5S6 A0A182S8Q9 A0A182YF53 A0A182XFE3 A0A182L7T6 A0A182J1F9 A0A182NG33 A0A084VTJ6 A0A182YF54 A0A1Q3EV39 A0A0N8DGW9 A0A0N8EED2 A0A0P5Y961 A0A162Q098 A0A0P5I633 A0A182GM26 B0WKK0 E9GJ60 A0A0P4ZMN0 A0A0P5G0R9 A0A182MTG7 A0A0P6DHJ3 A0A0P5ULU2 A0A0P5SG58 A0A0P5RV48 A0A0N8EE02 A0A0P5LZK4 A0A0P5GUL0 A0A0P5BQS1 Q17CY5 A0A1S4F7F6 A0A0P4ZXK5 A0A0P5S3L9 A0A0P5M7P6 A0A0P5IEW1 A0A0N7ZV01 A0A0P5TPL4 A0A162DAW2 A0A0P5EVV2 A0A0P5AI81 A0A182P439 A0A0P6JNR6 A0A0N7ZU83 A0A0P5BVJ2 A0A0P6EWY1
EC Number
1.11.1.7
Pubmed
EMBL
LC378389
BBD52153.1
BABH01032455
ODYU01008296
SOQ51753.1
ODYU01000151
+ More
SOQ34434.1 KZ149911 PZC78137.1 NWSH01000659 PCG74967.1 KQ459579 KPI99509.1 KQ459280 KPJ02047.1 KQ460542 KPJ14147.1 KQ971312 KYB29080.1 GEZM01033908 GEZM01033907 GEZM01033906 JAV84050.1 CH954181 EDV48685.1 CM000160 EDW95818.2 CM000364 EDX12533.1 AF238306 AE014297 AY119616 AAF78217.1 AAM50270.1 AAN13751.2 CH480827 EDW44698.1 KB632098 ERL88791.1 APGK01023128 KB740465 ENN80594.1 CH379064 EDY72527.1 OUUW01000015 SPP88579.1 GDHF01016115 JAI36199.1 CH964251 EDW83073.1 GAKP01010121 JAC48831.1 CH902623 EDV30170.1 AJWK01010594 AJWK01010595 AJWK01010596 AJWK01010597 AJWK01010598 AJWK01010599 AJWK01010600 AJWK01010601 AJWK01010602 CVRI01000002 CRK86945.1 JRES01001516 KNC22326.1 GAMC01000396 JAC06160.1 GBXI01016532 GBXI01003411 JAC97759.1 JAD10881.1 GFDF01000556 JAV13528.1 GAKP01010120 JAC48832.1 GFDF01000557 JAV13527.1 CP012526 ALC46682.1 GANO01002592 JAB57279.1 UFQS01000524 UFQT01000524 SSX04594.1 SSX24957.1 CH940652 EDW59877.2 CH916374 EDV91128.1 CH933806 EDW13884.2 AJVK01010081 AJVK01010082 AJVK01010083 AJVK01010084 AJVK01010085 AJVK01010086 AJVK01010087 AJVK01010088 AJVK01010089 AXCN02001195 GAMD01002595 JAA98995.1 AXCM01009432 ADMH02001557 ETN62073.1 IACF01005278 LAB70864.1 APCN01002644 AAAB01008817 AY752905 AAV30079.1 ATLV01016358 KE525079 KFB41290.1 GFDL01015873 JAV19172.1 GDIP01034895 JAM68820.1 GDIQ01035012 JAN59725.1 GDIP01062691 JAM41024.1 LRGB01000389 KZS19289.1 GDIQ01217481 JAK34244.1 JXUM01073690 JXUM01073691 JXUM01073692 KQ562789 KXJ75129.1 DS231973 EDS29894.1 GL732547 EFX80390.1 GDIP01210375 JAJ13027.1 GDIQ01248330 JAK03395.1 GDIQ01077337 JAN17400.1 GDIP01111182 JAL92532.1 GDIQ01096228 JAL55498.1 GDIQ01096229 JAL55497.1 GDIQ01036052 JAN58685.1 GDIQ01173558 JAK78167.1 GDIQ01239538 JAK12187.1 GDIP01181228 JAJ42174.1 CH477302 EAT44220.2 GDIP01209930 JAJ13472.1 GDIQ01110023 JAL41703.1 GDIQ01171501 JAK80224.1 GDIQ01215343 JAK36382.1 GDIP01209929 JAJ13473.1 GDIP01123736 GDIP01071343 JAL79978.1 LRGB01002076 KZS09294.1 GDIQ01266458 JAJ85266.1 GDIP01212074 JAJ11328.1 GDIQ01013552 JAN81185.1 GDIP01212073 GDIQ01187448 GDIQ01123724 JAJ11329.1 JAK64277.1 GDIP01179325 JAJ44077.1 GDIQ01069489 JAN25248.1
SOQ34434.1 KZ149911 PZC78137.1 NWSH01000659 PCG74967.1 KQ459579 KPI99509.1 KQ459280 KPJ02047.1 KQ460542 KPJ14147.1 KQ971312 KYB29080.1 GEZM01033908 GEZM01033907 GEZM01033906 JAV84050.1 CH954181 EDV48685.1 CM000160 EDW95818.2 CM000364 EDX12533.1 AF238306 AE014297 AY119616 AAF78217.1 AAM50270.1 AAN13751.2 CH480827 EDW44698.1 KB632098 ERL88791.1 APGK01023128 KB740465 ENN80594.1 CH379064 EDY72527.1 OUUW01000015 SPP88579.1 GDHF01016115 JAI36199.1 CH964251 EDW83073.1 GAKP01010121 JAC48831.1 CH902623 EDV30170.1 AJWK01010594 AJWK01010595 AJWK01010596 AJWK01010597 AJWK01010598 AJWK01010599 AJWK01010600 AJWK01010601 AJWK01010602 CVRI01000002 CRK86945.1 JRES01001516 KNC22326.1 GAMC01000396 JAC06160.1 GBXI01016532 GBXI01003411 JAC97759.1 JAD10881.1 GFDF01000556 JAV13528.1 GAKP01010120 JAC48832.1 GFDF01000557 JAV13527.1 CP012526 ALC46682.1 GANO01002592 JAB57279.1 UFQS01000524 UFQT01000524 SSX04594.1 SSX24957.1 CH940652 EDW59877.2 CH916374 EDV91128.1 CH933806 EDW13884.2 AJVK01010081 AJVK01010082 AJVK01010083 AJVK01010084 AJVK01010085 AJVK01010086 AJVK01010087 AJVK01010088 AJVK01010089 AXCN02001195 GAMD01002595 JAA98995.1 AXCM01009432 ADMH02001557 ETN62073.1 IACF01005278 LAB70864.1 APCN01002644 AAAB01008817 AY752905 AAV30079.1 ATLV01016358 KE525079 KFB41290.1 GFDL01015873 JAV19172.1 GDIP01034895 JAM68820.1 GDIQ01035012 JAN59725.1 GDIP01062691 JAM41024.1 LRGB01000389 KZS19289.1 GDIQ01217481 JAK34244.1 JXUM01073690 JXUM01073691 JXUM01073692 KQ562789 KXJ75129.1 DS231973 EDS29894.1 GL732547 EFX80390.1 GDIP01210375 JAJ13027.1 GDIQ01248330 JAK03395.1 GDIQ01077337 JAN17400.1 GDIP01111182 JAL92532.1 GDIQ01096228 JAL55498.1 GDIQ01096229 JAL55497.1 GDIQ01036052 JAN58685.1 GDIQ01173558 JAK78167.1 GDIQ01239538 JAK12187.1 GDIP01181228 JAJ42174.1 CH477302 EAT44220.2 GDIP01209930 JAJ13472.1 GDIQ01110023 JAL41703.1 GDIQ01171501 JAK80224.1 GDIQ01215343 JAK36382.1 GDIP01209929 JAJ13473.1 GDIP01123736 GDIP01071343 JAL79978.1 LRGB01002076 KZS09294.1 GDIQ01266458 JAJ85266.1 GDIP01212074 JAJ11328.1 GDIQ01013552 JAN81185.1 GDIP01212073 GDIQ01187448 GDIQ01123724 JAJ11329.1 JAK64277.1 GDIP01179325 JAJ44077.1 GDIQ01069489 JAN25248.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007266
UP000192221
+ More
UP000008711 UP000002282 UP000000304 UP000000803 UP000001292 UP000030742 UP000019118 UP000001819 UP000268350 UP000007798 UP000007801 UP000092461 UP000183832 UP000037069 UP000095300 UP000095301 UP000092553 UP000008792 UP000001070 UP000009192 UP000092462 UP000075881 UP000075886 UP000075883 UP000000673 UP000075900 UP000075920 UP000075840 UP000007062 UP000075902 UP000075885 UP000075903 UP000075901 UP000076408 UP000076407 UP000075882 UP000075880 UP000075884 UP000030765 UP000076858 UP000069940 UP000249989 UP000002320 UP000000305 UP000008820
UP000008711 UP000002282 UP000000304 UP000000803 UP000001292 UP000030742 UP000019118 UP000001819 UP000268350 UP000007798 UP000007801 UP000092461 UP000183832 UP000037069 UP000095300 UP000095301 UP000092553 UP000008792 UP000001070 UP000009192 UP000092462 UP000075881 UP000075886 UP000075883 UP000000673 UP000075900 UP000075920 UP000075840 UP000007062 UP000075902 UP000075885 UP000075903 UP000075901 UP000076408 UP000076407 UP000075882 UP000075880 UP000075884 UP000030765 UP000076858 UP000069940 UP000249989 UP000002320 UP000000305 UP000008820
Interpro
Gene 3D
ProteinModelPortal
A0A348B065
H9JVS7
A0A2H1WFB0
A0A2H1V2D9
A0A2W1BY28
A0A2A4JSF2
+ More
A0A194Q3C2 A0A0N0P9N7 A0A0N1PI15 A0A139WMA7 A0A1Y1MHD3 A0A1W4V5J1 B3P0H0 B4PKH9 B4QV50 Q9VEG6 B4IBE0 U4UDX4 N6TJI6 B5DM11 A0A3B0KPN2 A0A0K8VB63 B4NFK6 A0A034VZV8 B3MTN9 A0A1B0CFZ9 A0A1J1HL34 A0A0L0BQN4 A0A1I8PRJ4 W8CED3 A0A0A1WGZ5 A0A1L8E461 A0A034W3E1 A0A1L8E467 A0A1I8MYK6 A0A0M4F596 U5ES00 A0A336KIB1 B4M5G6 B4JUF8 B4KDQ0 A0A1B0DRI5 A0A182JWM4 A0A182QE50 T1DNT2 A0A182M8J6 W5JFF7 A0A182RSK4 A0A182VX07 A0A2P2IA20 A0A182I859 Q7QH73 A0A182UH95 A0A182P440 A0A182V5S6 A0A182S8Q9 A0A182YF53 A0A182XFE3 A0A182L7T6 A0A182J1F9 A0A182NG33 A0A084VTJ6 A0A182YF54 A0A1Q3EV39 A0A0N8DGW9 A0A0N8EED2 A0A0P5Y961 A0A162Q098 A0A0P5I633 A0A182GM26 B0WKK0 E9GJ60 A0A0P4ZMN0 A0A0P5G0R9 A0A182MTG7 A0A0P6DHJ3 A0A0P5ULU2 A0A0P5SG58 A0A0P5RV48 A0A0N8EE02 A0A0P5LZK4 A0A0P5GUL0 A0A0P5BQS1 Q17CY5 A0A1S4F7F6 A0A0P4ZXK5 A0A0P5S3L9 A0A0P5M7P6 A0A0P5IEW1 A0A0N7ZV01 A0A0P5TPL4 A0A162DAW2 A0A0P5EVV2 A0A0P5AI81 A0A182P439 A0A0P6JNR6 A0A0N7ZU83 A0A0P5BVJ2 A0A0P6EWY1
A0A194Q3C2 A0A0N0P9N7 A0A0N1PI15 A0A139WMA7 A0A1Y1MHD3 A0A1W4V5J1 B3P0H0 B4PKH9 B4QV50 Q9VEG6 B4IBE0 U4UDX4 N6TJI6 B5DM11 A0A3B0KPN2 A0A0K8VB63 B4NFK6 A0A034VZV8 B3MTN9 A0A1B0CFZ9 A0A1J1HL34 A0A0L0BQN4 A0A1I8PRJ4 W8CED3 A0A0A1WGZ5 A0A1L8E461 A0A034W3E1 A0A1L8E467 A0A1I8MYK6 A0A0M4F596 U5ES00 A0A336KIB1 B4M5G6 B4JUF8 B4KDQ0 A0A1B0DRI5 A0A182JWM4 A0A182QE50 T1DNT2 A0A182M8J6 W5JFF7 A0A182RSK4 A0A182VX07 A0A2P2IA20 A0A182I859 Q7QH73 A0A182UH95 A0A182P440 A0A182V5S6 A0A182S8Q9 A0A182YF53 A0A182XFE3 A0A182L7T6 A0A182J1F9 A0A182NG33 A0A084VTJ6 A0A182YF54 A0A1Q3EV39 A0A0N8DGW9 A0A0N8EED2 A0A0P5Y961 A0A162Q098 A0A0P5I633 A0A182GM26 B0WKK0 E9GJ60 A0A0P4ZMN0 A0A0P5G0R9 A0A182MTG7 A0A0P6DHJ3 A0A0P5ULU2 A0A0P5SG58 A0A0P5RV48 A0A0N8EE02 A0A0P5LZK4 A0A0P5GUL0 A0A0P5BQS1 Q17CY5 A0A1S4F7F6 A0A0P4ZXK5 A0A0P5S3L9 A0A0P5M7P6 A0A0P5IEW1 A0A0N7ZV01 A0A0P5TPL4 A0A162DAW2 A0A0P5EVV2 A0A0P5AI81 A0A182P439 A0A0P6JNR6 A0A0N7ZU83 A0A0P5BVJ2 A0A0P6EWY1
PDB
5HPW
E-value=1.03473e-71,
Score=689
Ontologies
GO
PANTHER
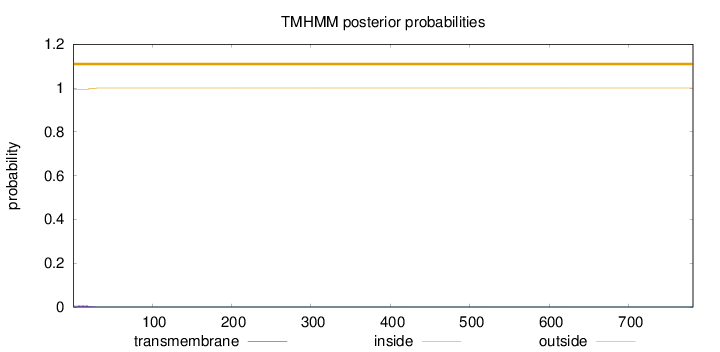
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 24,
Likelihood: 0.995348

Length:
781
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11471
Exp number, first 60 AAs:
0.11179
Total prob of N-in:
0.00590
outside
1 - 781
Population Genetic Test Statistics
Pi
268.984047
Theta
170.048774
Tajima's D
1.480283
CLR
0.493008
CSRT
0.777061146942653
Interpretation
Uncertain
