Gene
KWMTBOMO03206
Annotation
PREDICTED:_synaptic_vesicle_glycoprotein_2B-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.299
Sequence
CDS
ATGAGGGAACCAGGCCAGGGAATAGGTGGTGTCGCGAGTGCGTTCTTCTGGGGTATTGTGGGCGATGTCTTCGGCAGGAAGAGCACCCTAAGCTTGACATTGCTCCTGGACGCAGCGATAACTTTAGCCCAGAGCACCGTTCCAGACTACAGACTTCTGCTGGCAGCTAGAACCATCAATGGGTTCATTATAGGAGGTCCAGCTACACTGGTCTTTACGTATTTATCAGACCTAGTTGGAGTTAAGAAGAGACAATTCTATCTTAGCATGGTGGGAATGAGTTTTGTTGCGGCCTGGTTACTTTTGCCTGGATTCACTAAATAG
Protein
MREPGQGIGGVASAFFWGIVGDVFGRKSTLSLTLLLDAAITLAQSTVPDYRLLLAARTINGFIIGGPATLVFTYLSDLVGVKKRQFYLSMVGMSFVAAWLLLPGFTK
Summary
Uniprot
A0A2W1BZ69
A0A2A4JP45
A0A2H1V0P6
A0A0N1IEI8
A0A212EXZ1
A0A194Q7X5
+ More
A0A1B6DD61 A0A0C9RT66 A0A0C9PZZ9 A0A151XCA7 A0A0L7QLM7 A0A0M9A095 A0A310SDB3 A0A154NYA8 A0A195B1S0 A0A158NLE1 E2ATB0 E2BMY5 A0A088AG68 F4WW57 A0A2A3EPR0 A0A195CY83 A0A1W4WFM4 A0A195F2P9 A0A3L8D687 A0A026WID1 E0VD11 A0A195E6G9 A0A0T6B5Y4 E2BMY6 W8C4Z8 R4WRQ1 A0A1I8PT34 A0A1I8PT04 A0A087XTG0 A0A146LC37 A0A3B4DUA9 W5L2M0 A0A1L8H0Z8 T1PAI7 A0A1I8M6J6 A0A3B4VPU2 A0A3B5MH50 E0W3U1 A0A1A7XXM3 A0A0K8SJZ1 A0A3P9Q0K1 A0A096M6X8 A0A146W9Q5 M4AWZ8 A0A3B3VJ41 F6QL33 A0A0A9WZX1 A0A3B3X3B6 A0A3B5AD85 A0A2U9BA84 A0A1A7Y1V0 A0A146P368 A0A0K8VAX0 H2VCQ2 A2CF25 A0A1S3KZJ5 A0A3P8S2E0 A0A286Y8C7 A0A1A7ZLP8 A0A1Y1LT93 B4LZI9 A0A1L8HRE3 A0A1A8ETI5 U4UDT0 J3JVU6 A0A2D0PPV4 G1N043 G1N9R7 A0A3P9JS84 M3W047 A0A3P9JT06 A0A3B3I7N0 H2LA02 A0A3P9BH25 A0A0K8VDZ3 A0A3P9KX51 A0A3P8ZMG2 F6W539 A0A1L8I228 A0A3P8R9Z4 A0A3P9BH20 A0A3P8VU01 A0A3B4FCV0 A0A0A1XHP3 N6U3Q5 H3DB15 U4U2W7 A0A3B3CD39 A0A3B4AWN8 H3D7I9 Q4S265 A0A3B4AVC8 B4LAH0
A0A1B6DD61 A0A0C9RT66 A0A0C9PZZ9 A0A151XCA7 A0A0L7QLM7 A0A0M9A095 A0A310SDB3 A0A154NYA8 A0A195B1S0 A0A158NLE1 E2ATB0 E2BMY5 A0A088AG68 F4WW57 A0A2A3EPR0 A0A195CY83 A0A1W4WFM4 A0A195F2P9 A0A3L8D687 A0A026WID1 E0VD11 A0A195E6G9 A0A0T6B5Y4 E2BMY6 W8C4Z8 R4WRQ1 A0A1I8PT34 A0A1I8PT04 A0A087XTG0 A0A146LC37 A0A3B4DUA9 W5L2M0 A0A1L8H0Z8 T1PAI7 A0A1I8M6J6 A0A3B4VPU2 A0A3B5MH50 E0W3U1 A0A1A7XXM3 A0A0K8SJZ1 A0A3P9Q0K1 A0A096M6X8 A0A146W9Q5 M4AWZ8 A0A3B3VJ41 F6QL33 A0A0A9WZX1 A0A3B3X3B6 A0A3B5AD85 A0A2U9BA84 A0A1A7Y1V0 A0A146P368 A0A0K8VAX0 H2VCQ2 A2CF25 A0A1S3KZJ5 A0A3P8S2E0 A0A286Y8C7 A0A1A7ZLP8 A0A1Y1LT93 B4LZI9 A0A1L8HRE3 A0A1A8ETI5 U4UDT0 J3JVU6 A0A2D0PPV4 G1N043 G1N9R7 A0A3P9JS84 M3W047 A0A3P9JT06 A0A3B3I7N0 H2LA02 A0A3P9BH25 A0A0K8VDZ3 A0A3P9KX51 A0A3P8ZMG2 F6W539 A0A1L8I228 A0A3P8R9Z4 A0A3P9BH20 A0A3P8VU01 A0A3B4FCV0 A0A0A1XHP3 N6U3Q5 H3DB15 U4U2W7 A0A3B3CD39 A0A3B4AWN8 H3D7I9 Q4S265 A0A3B4AVC8 B4LAH0
Pubmed
28756777
26354079
22118469
21347285
20798317
21719571
+ More
30249741 24508170 20566863 24495485 23691247 26823975 25329095 27762356 25315136 23542700 20431018 25401762 21551351 23594743 28004739 17994087 23537049 22516182 20838655 17554307 17975172 25186727 25069045 24487278 25830018 15496914 29451363 25463417
30249741 24508170 20566863 24495485 23691247 26823975 25329095 27762356 25315136 23542700 20431018 25401762 21551351 23594743 28004739 17994087 23537049 22516182 20838655 17554307 17975172 25186727 25069045 24487278 25830018 15496914 29451363 25463417
EMBL
KZ149911
PZC78140.1
NWSH01000857
PCG73827.1
ODYU01000151
SOQ34433.1
+ More
KQ460542 KPJ14148.1 AGBW02011638 OWR46363.1 KQ459579 KPI99510.1 GEDC01013690 JAS23608.1 GBYB01011890 JAG81657.1 GBYB01007148 JAG76915.1 KQ982314 KYQ58026.1 KQ414914 KOC59533.1 KQ435801 KOX73253.1 KQ775195 OAD52273.1 KQ434773 KZC04008.1 KQ976662 KYM78421.1 ADTU01019324 GL442545 EFN63355.1 GL449382 EFN82889.1 GL888404 EGI61528.1 KZ288197 PBC33718.1 KQ977115 KYN05613.1 KQ981864 KYN34369.1 QOIP01000012 RLU15749.1 KK107185 EZA55807.1 DS235070 EEB11267.1 KQ979568 KYN20768.1 LJIG01009608 KRT82740.1 EFN82890.1 GAMC01009291 JAB97264.1 AK418431 BAN21601.1 AYCK01003184 AYCK01003185 AYCK01003186 GDHC01014499 JAQ04130.1 CM004470 OCT89763.1 KA645786 AFP60415.1 DS235883 EEB20297.1 HADX01000655 SBP22887.1 GBRD01012207 JAG53617.1 GCES01059530 JAR26793.1 AAMC01021764 GBHO01030315 JAG13289.1 CP026246 AWP00672.1 HADX01002170 SBP24402.1 GCES01147994 JAQ38328.1 GDHF01016291 JAI36023.1 BX294131 CR854913 HADY01004465 HAEJ01005941 SBP42950.1 GEZM01047127 JAV76924.1 CH940650 EDW67128.1 CM004467 OCT98658.1 HAEB01003650 SBQ50177.1 KB632277 ERL91212.1 BT127364 AEE62326.1 AANG04001174 GDHF01015150 JAI37164.1 AAMC01037688 AAMC01037689 AAMC01037690 AAMC01037691 AAMC01037692 CM004466 OCU02410.1 GBXI01003857 JAD10435.1 APGK01043766 APGK01043767 KB741017 ENN75251.1 KB631644 ERL84936.1 CAAE01014764 CAG05267.1 CH934611 EDW17078.2
KQ460542 KPJ14148.1 AGBW02011638 OWR46363.1 KQ459579 KPI99510.1 GEDC01013690 JAS23608.1 GBYB01011890 JAG81657.1 GBYB01007148 JAG76915.1 KQ982314 KYQ58026.1 KQ414914 KOC59533.1 KQ435801 KOX73253.1 KQ775195 OAD52273.1 KQ434773 KZC04008.1 KQ976662 KYM78421.1 ADTU01019324 GL442545 EFN63355.1 GL449382 EFN82889.1 GL888404 EGI61528.1 KZ288197 PBC33718.1 KQ977115 KYN05613.1 KQ981864 KYN34369.1 QOIP01000012 RLU15749.1 KK107185 EZA55807.1 DS235070 EEB11267.1 KQ979568 KYN20768.1 LJIG01009608 KRT82740.1 EFN82890.1 GAMC01009291 JAB97264.1 AK418431 BAN21601.1 AYCK01003184 AYCK01003185 AYCK01003186 GDHC01014499 JAQ04130.1 CM004470 OCT89763.1 KA645786 AFP60415.1 DS235883 EEB20297.1 HADX01000655 SBP22887.1 GBRD01012207 JAG53617.1 GCES01059530 JAR26793.1 AAMC01021764 GBHO01030315 JAG13289.1 CP026246 AWP00672.1 HADX01002170 SBP24402.1 GCES01147994 JAQ38328.1 GDHF01016291 JAI36023.1 BX294131 CR854913 HADY01004465 HAEJ01005941 SBP42950.1 GEZM01047127 JAV76924.1 CH940650 EDW67128.1 CM004467 OCT98658.1 HAEB01003650 SBQ50177.1 KB632277 ERL91212.1 BT127364 AEE62326.1 AANG04001174 GDHF01015150 JAI37164.1 AAMC01037688 AAMC01037689 AAMC01037690 AAMC01037691 AAMC01037692 CM004466 OCU02410.1 GBXI01003857 JAD10435.1 APGK01043766 APGK01043767 KB741017 ENN75251.1 KB631644 ERL84936.1 CAAE01014764 CAG05267.1 CH934611 EDW17078.2
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000075809
UP000053825
+ More
UP000053105 UP000076502 UP000078540 UP000005205 UP000000311 UP000008237 UP000005203 UP000007755 UP000242457 UP000078542 UP000192223 UP000078541 UP000279307 UP000053097 UP000009046 UP000078492 UP000095300 UP000028760 UP000261440 UP000018467 UP000186698 UP000095301 UP000261420 UP000261380 UP000242638 UP000002852 UP000261500 UP000008143 UP000261480 UP000261400 UP000246464 UP000005226 UP000000437 UP000087266 UP000265080 UP000008792 UP000030742 UP000221080 UP000001645 UP000265200 UP000011712 UP000001038 UP000265160 UP000265180 UP000265140 UP000265100 UP000265120 UP000261460 UP000019118 UP000007303 UP000261560 UP000261520 UP000009192
UP000053105 UP000076502 UP000078540 UP000005205 UP000000311 UP000008237 UP000005203 UP000007755 UP000242457 UP000078542 UP000192223 UP000078541 UP000279307 UP000053097 UP000009046 UP000078492 UP000095300 UP000028760 UP000261440 UP000018467 UP000186698 UP000095301 UP000261420 UP000261380 UP000242638 UP000002852 UP000261500 UP000008143 UP000261480 UP000261400 UP000246464 UP000005226 UP000000437 UP000087266 UP000265080 UP000008792 UP000030742 UP000221080 UP000001645 UP000265200 UP000011712 UP000001038 UP000265160 UP000265180 UP000265140 UP000265100 UP000265120 UP000261460 UP000019118 UP000007303 UP000261560 UP000261520 UP000009192
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2W1BZ69
A0A2A4JP45
A0A2H1V0P6
A0A0N1IEI8
A0A212EXZ1
A0A194Q7X5
+ More
A0A1B6DD61 A0A0C9RT66 A0A0C9PZZ9 A0A151XCA7 A0A0L7QLM7 A0A0M9A095 A0A310SDB3 A0A154NYA8 A0A195B1S0 A0A158NLE1 E2ATB0 E2BMY5 A0A088AG68 F4WW57 A0A2A3EPR0 A0A195CY83 A0A1W4WFM4 A0A195F2P9 A0A3L8D687 A0A026WID1 E0VD11 A0A195E6G9 A0A0T6B5Y4 E2BMY6 W8C4Z8 R4WRQ1 A0A1I8PT34 A0A1I8PT04 A0A087XTG0 A0A146LC37 A0A3B4DUA9 W5L2M0 A0A1L8H0Z8 T1PAI7 A0A1I8M6J6 A0A3B4VPU2 A0A3B5MH50 E0W3U1 A0A1A7XXM3 A0A0K8SJZ1 A0A3P9Q0K1 A0A096M6X8 A0A146W9Q5 M4AWZ8 A0A3B3VJ41 F6QL33 A0A0A9WZX1 A0A3B3X3B6 A0A3B5AD85 A0A2U9BA84 A0A1A7Y1V0 A0A146P368 A0A0K8VAX0 H2VCQ2 A2CF25 A0A1S3KZJ5 A0A3P8S2E0 A0A286Y8C7 A0A1A7ZLP8 A0A1Y1LT93 B4LZI9 A0A1L8HRE3 A0A1A8ETI5 U4UDT0 J3JVU6 A0A2D0PPV4 G1N043 G1N9R7 A0A3P9JS84 M3W047 A0A3P9JT06 A0A3B3I7N0 H2LA02 A0A3P9BH25 A0A0K8VDZ3 A0A3P9KX51 A0A3P8ZMG2 F6W539 A0A1L8I228 A0A3P8R9Z4 A0A3P9BH20 A0A3P8VU01 A0A3B4FCV0 A0A0A1XHP3 N6U3Q5 H3DB15 U4U2W7 A0A3B3CD39 A0A3B4AWN8 H3D7I9 Q4S265 A0A3B4AVC8 B4LAH0
A0A1B6DD61 A0A0C9RT66 A0A0C9PZZ9 A0A151XCA7 A0A0L7QLM7 A0A0M9A095 A0A310SDB3 A0A154NYA8 A0A195B1S0 A0A158NLE1 E2ATB0 E2BMY5 A0A088AG68 F4WW57 A0A2A3EPR0 A0A195CY83 A0A1W4WFM4 A0A195F2P9 A0A3L8D687 A0A026WID1 E0VD11 A0A195E6G9 A0A0T6B5Y4 E2BMY6 W8C4Z8 R4WRQ1 A0A1I8PT34 A0A1I8PT04 A0A087XTG0 A0A146LC37 A0A3B4DUA9 W5L2M0 A0A1L8H0Z8 T1PAI7 A0A1I8M6J6 A0A3B4VPU2 A0A3B5MH50 E0W3U1 A0A1A7XXM3 A0A0K8SJZ1 A0A3P9Q0K1 A0A096M6X8 A0A146W9Q5 M4AWZ8 A0A3B3VJ41 F6QL33 A0A0A9WZX1 A0A3B3X3B6 A0A3B5AD85 A0A2U9BA84 A0A1A7Y1V0 A0A146P368 A0A0K8VAX0 H2VCQ2 A2CF25 A0A1S3KZJ5 A0A3P8S2E0 A0A286Y8C7 A0A1A7ZLP8 A0A1Y1LT93 B4LZI9 A0A1L8HRE3 A0A1A8ETI5 U4UDT0 J3JVU6 A0A2D0PPV4 G1N043 G1N9R7 A0A3P9JS84 M3W047 A0A3P9JT06 A0A3B3I7N0 H2LA02 A0A3P9BH25 A0A0K8VDZ3 A0A3P9KX51 A0A3P8ZMG2 F6W539 A0A1L8I228 A0A3P8R9Z4 A0A3P9BH20 A0A3P8VU01 A0A3B4FCV0 A0A0A1XHP3 N6U3Q5 H3DB15 U4U2W7 A0A3B3CD39 A0A3B4AWN8 H3D7I9 Q4S265 A0A3B4AVC8 B4LAH0
Ontologies
GO
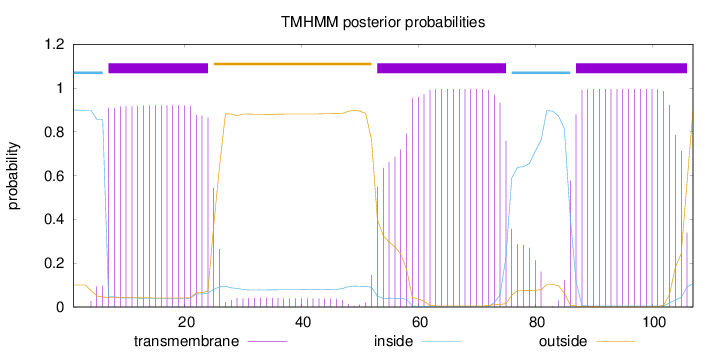
Topology
Length:
107
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
59.78817
Exp number, first 60 AAs:
24.36475
Total prob of N-in:
0.89916
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 52
TMhelix
53 - 75
inside
76 - 86
TMhelix
87 - 106
outside
107 - 107
Population Genetic Test Statistics
Pi
177.7318
Theta
146.739819
Tajima's D
-0.220288
CLR
0.000012
CSRT
0.311184440777961
Interpretation
Uncertain
