Gene
KWMTBOMO03204
Pre Gene Modal
BGIBMGA013574
Annotation
PREDICTED:_synaptic_vesicle_glycoprotein_2B_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.491
Sequence
CDS
ATGTTATTAATGACCTGCCTATTGTTATTAGCACCAGCTGTTTCTGGAGTTTTAACCTATCCCACGTTGAAGTTAGACTTCGCCACGGACCTATCAACTATAGGAGTCGTCTACAAGCCTTGGAGGTTGCTCATCATCGTACTGGCTCTTCCACTAGGAATGGGTGCATTAGCCATGTTTTTCTTTTACGAATCTCCAAAATTCCTTTTCAACTGTGGAAGAAAAGACGAAGCTATGGAGGTGCTGAAGGGAATTTACGCTATCAACCATAGAACCTCGAAAAATAACTACAAGATTCGGTTAATGAAAATAGAAAGAAAAACAGAAACGAAATCGGCATCTATCTGGCGCGCCGTTTACGAACAGAGCGCTCCCCTATTTCGCGAGCCGTATCTCCTGAGAAGCCTGCAGCTGTTCTACATTGTTTCCGTGGCGTACATCACCAATAACAGCTTCTTGATCTGGCTCCCGCACATCATGGAGCAAGTTAGGAAATCCTTACAAGCTAACGGCACTAGAACCGGTAACGTCTGTGAACTGATTCATACGGAAGCAACAGGAAATCTTTCCTCAAGCGACTCCTCGGAGAACTGCTTGGGAACCGTGGAAGATAACGTGGTATTCACTCTCATAATATCCCAGACCATATTTTCCCTTTTAAACTTTGTATTATCTTATCTACCCAACCACCGAAGGCCGGTGCTCCTCACAGTTCTGTCCGTCTCAGCCGTGAGCGGGGTCCTGATCAACTTGACGCCGGAGACAATATCCAGTGTGATATTCTTCATGATGTTCACTTGCACTTGCTTGGGTATGGGGATCATGGCGTCCTATTTTGTGGACCTGTACCCAACATCTCACAGGGGAATGGTGACGTGCCTAAGTATCATGGTGGGCAGGAGTTCGGCATTCATGGGAATCAATTTAGTTGGACACCTCATATTCAACTACTGTCAGTTAACATTTTATCTCTGGTCTTTGTTGGTTTTAACCAGTGTCGTGGCAGCATGGTTTCTTCCCCCAGATCGACATGAGAAGCTGAAAACGTCTAACTGA
Protein
MLLMTCLLLLAPAVSGVLTYPTLKLDFATDLSTIGVVYKPWRLLIIVLALPLGMGALAMFFFYESPKFLFNCGRKDEAMEVLKGIYAINHRTSKNNYKIRLMKIERKTETKSASIWRAVYEQSAPLFREPYLLRSLQLFYIVSVAYITNNSFLIWLPHIMEQVRKSLQANGTRTGNVCELIHTEATGNLSSSDSSENCLGTVEDNVVFTLIISQTIFSLLNFVLSYLPNHRRPVLLTVLSVSAVSGVLINLTPETISSVIFFMMFTCTCLGMGIMASYFVDLYPTSHRGMVTCLSIMVGRSSAFMGINLVGHLIFNYCQLTFYLWSLLVLTSVVAAWFLPPDRHEKLKTSN
Summary
Uniprot
H9JVL1
A0A2W1BUU8
A0A2H1V250
A0A194Q1T6
A0A212EY43
A0A2H1WI98
+ More
A0A2A4JL38 A0A0L7K4N0 A0A194Q2D2 H9JVM2 A0A0N1IG89 A0A2A4JAH3 A0A194Q800 A0A194Q1Q2 A0A2A4IYX6 A0A194Q1T1 A0A2H1WJU4 A0A2W1BVQ5 A0A0L7L435 A0A212FGQ8 A0A2H1V0R1 A0A212F5C2 A0A2H1VUC9 A0A2A4J999 A0A0N0PCK2 A0A2W1BGE8 H9JV72 A0A2H1W801 S4Q006 A0A0N1I9N3 A0A2A4JT62 A0A2H1WUV1 A0A2W1BZI9 A0A2H1VEB4 A0A0L7LHZ8 A0A2H1VUK7 A0A2H1WU82 A0A2H1VVE4 A0A0N0PCN2 A0A2W1BWY0 H9JVL3 A0A2H1WGI5 A0A2A4JX40 A0A194Q1R5 A0A212EQC0 A0A0N1IPG1 H9JVR6 A0A2H1W7K7 A0A1Q3FNL7 A0A1Q3FNK2 A0A1I8P9Y1 B0W415 A0A2A4JKW1 A0A2A4JLN1 A0A1Q3FM89 A0A1Q3FLR3 A0A034VU45 T1GER7 A0A034VUT9 A0A2H1X0I9 A0A0K8UIB9 A0A1I8P1U8 A0A1I8P1R5 A0A1L8E785 W8B9U3 A0A1L8E7E8 A0A1L8E7J3 A0A0A1XE60 W8B5D0 A0A1I8MVX6 A0A1L8EFQ6 A0A1I8MVY3 A0A1L8EFY3 T1PCW5 A0A1L8EFU2 A0A212EZN8 A0A194PJI3 B0WKE4 A0A2A4JLT5 A0A0A1XC67 A0A1A9X005 A0A212EYD8 U5ETH9 A0A1I8MZK1 A0A182GKA8 A0A2A4K5Q7 A0A2H1WYY1 A0A182GYQ4 A0A2W1BLZ7 A0A0L0BUM3 A0A2A4JLI7 A0A1A9WGN7
A0A2A4JL38 A0A0L7K4N0 A0A194Q2D2 H9JVM2 A0A0N1IG89 A0A2A4JAH3 A0A194Q800 A0A194Q1Q2 A0A2A4IYX6 A0A194Q1T1 A0A2H1WJU4 A0A2W1BVQ5 A0A0L7L435 A0A212FGQ8 A0A2H1V0R1 A0A212F5C2 A0A2H1VUC9 A0A2A4J999 A0A0N0PCK2 A0A2W1BGE8 H9JV72 A0A2H1W801 S4Q006 A0A0N1I9N3 A0A2A4JT62 A0A2H1WUV1 A0A2W1BZI9 A0A2H1VEB4 A0A0L7LHZ8 A0A2H1VUK7 A0A2H1WU82 A0A2H1VVE4 A0A0N0PCN2 A0A2W1BWY0 H9JVL3 A0A2H1WGI5 A0A2A4JX40 A0A194Q1R5 A0A212EQC0 A0A0N1IPG1 H9JVR6 A0A2H1W7K7 A0A1Q3FNL7 A0A1Q3FNK2 A0A1I8P9Y1 B0W415 A0A2A4JKW1 A0A2A4JLN1 A0A1Q3FM89 A0A1Q3FLR3 A0A034VU45 T1GER7 A0A034VUT9 A0A2H1X0I9 A0A0K8UIB9 A0A1I8P1U8 A0A1I8P1R5 A0A1L8E785 W8B9U3 A0A1L8E7E8 A0A1L8E7J3 A0A0A1XE60 W8B5D0 A0A1I8MVX6 A0A1L8EFQ6 A0A1I8MVY3 A0A1L8EFY3 T1PCW5 A0A1L8EFU2 A0A212EZN8 A0A194PJI3 B0WKE4 A0A2A4JLT5 A0A0A1XC67 A0A1A9X005 A0A212EYD8 U5ETH9 A0A1I8MZK1 A0A182GKA8 A0A2A4K5Q7 A0A2H1WYY1 A0A182GYQ4 A0A2W1BLZ7 A0A0L0BUM3 A0A2A4JLI7 A0A1A9WGN7
Pubmed
EMBL
BABH01032458
KZ149911
PZC78141.1
ODYU01000151
SOQ34432.1
KQ459579
+ More
KPI99511.1 AGBW02011638 OWR46364.1 ODYU01008516 SOQ52184.1 NWSH01001089 PCG72681.1 JTDY01010294 KOB58258.1 KPI99493.1 BABH01032396 BABH01032397 BABH01032398 BABH01032399 KQ460542 KPJ14129.1 NWSH01002380 PCG68430.1 KPI99535.1 KPI99507.1 NWSH01005206 PCG64352.1 KPI99506.1 ODYU01009141 SOQ53353.1 KZ149979 PZC75843.1 JTDY01003063 KOB70227.1 AGBW02008612 OWR52914.1 SOQ34435.1 AGBW02010212 OWR48924.1 ODYU01004504 SOQ44451.1 PCG68429.1 KPJ14144.1 KZ150202 PZC72277.1 BABH01037389 ODYU01006861 SOQ49086.1 GAIX01000623 JAA91937.1 KPJ14146.1 NWSH01000659 PCG74966.1 ODYU01011202 SOQ56787.1 PZC78136.1 ODYU01001846 SOQ38614.1 JTDY01001109 KOB74851.1 ODYU01004537 SOQ44529.1 ODYU01011059 SOQ56557.1 SOQ44452.1 KPJ14131.1 PZC78134.1 BABH01032453 SOQ52183.1 NWSH01000457 PCG76268.1 KPI99491.1 AGBW02013272 OWR43679.1 KPJ14127.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 SOQ49085.1 GFDL01005969 JAV29076.1 GFDL01005870 JAV29175.1 DS231834 EDS32608.1 PCG72687.1 PCG72686.1 GFDL01006432 JAV28613.1 GFDL01006485 JAV28560.1 GAKP01013340 JAC45612.1 CAQQ02083380 CAQQ02083381 GAKP01013342 JAC45610.1 ODYU01012422 SOQ58716.1 GDHF01025885 JAI26429.1 GFDG01004236 JAV14563.1 GAMC01012742 JAB93813.1 GFDG01004235 JAV14564.1 GFDG01004234 JAV14565.1 GBXI01005544 JAD08748.1 GAMC01012743 JAB93812.1 GFDG01001242 JAV17557.1 GFDG01001245 JAV17554.1 KA646499 AFP61128.1 GFDG01001241 JAV17558.1 AGBW02011241 OWR46942.1 KQ459603 KPI92884.1 DS231971 EDS29763.1 PCG72678.1 GBXI01006239 JAD08053.1 AGBW02011531 OWR46513.1 GANO01004365 JAB55506.1 JXUM01069579 KQ562558 KXJ75630.1 NWSH01000098 PCG79585.1 ODYU01012071 SOQ58176.1 JXUM01021492 KQ560602 KXJ81676.1 KZ149975 PZC75948.1 JRES01001303 KNC23770.1 PCG72679.1
KPI99511.1 AGBW02011638 OWR46364.1 ODYU01008516 SOQ52184.1 NWSH01001089 PCG72681.1 JTDY01010294 KOB58258.1 KPI99493.1 BABH01032396 BABH01032397 BABH01032398 BABH01032399 KQ460542 KPJ14129.1 NWSH01002380 PCG68430.1 KPI99535.1 KPI99507.1 NWSH01005206 PCG64352.1 KPI99506.1 ODYU01009141 SOQ53353.1 KZ149979 PZC75843.1 JTDY01003063 KOB70227.1 AGBW02008612 OWR52914.1 SOQ34435.1 AGBW02010212 OWR48924.1 ODYU01004504 SOQ44451.1 PCG68429.1 KPJ14144.1 KZ150202 PZC72277.1 BABH01037389 ODYU01006861 SOQ49086.1 GAIX01000623 JAA91937.1 KPJ14146.1 NWSH01000659 PCG74966.1 ODYU01011202 SOQ56787.1 PZC78136.1 ODYU01001846 SOQ38614.1 JTDY01001109 KOB74851.1 ODYU01004537 SOQ44529.1 ODYU01011059 SOQ56557.1 SOQ44452.1 KPJ14131.1 PZC78134.1 BABH01032453 SOQ52183.1 NWSH01000457 PCG76268.1 KPI99491.1 AGBW02013272 OWR43679.1 KPJ14127.1 BABH01032418 BABH01032419 BABH01032420 BABH01032421 BABH01032422 BABH01032423 SOQ49085.1 GFDL01005969 JAV29076.1 GFDL01005870 JAV29175.1 DS231834 EDS32608.1 PCG72687.1 PCG72686.1 GFDL01006432 JAV28613.1 GFDL01006485 JAV28560.1 GAKP01013340 JAC45612.1 CAQQ02083380 CAQQ02083381 GAKP01013342 JAC45610.1 ODYU01012422 SOQ58716.1 GDHF01025885 JAI26429.1 GFDG01004236 JAV14563.1 GAMC01012742 JAB93813.1 GFDG01004235 JAV14564.1 GFDG01004234 JAV14565.1 GBXI01005544 JAD08748.1 GAMC01012743 JAB93812.1 GFDG01001242 JAV17557.1 GFDG01001245 JAV17554.1 KA646499 AFP61128.1 GFDG01001241 JAV17558.1 AGBW02011241 OWR46942.1 KQ459603 KPI92884.1 DS231971 EDS29763.1 PCG72678.1 GBXI01006239 JAD08053.1 AGBW02011531 OWR46513.1 GANO01004365 JAB55506.1 JXUM01069579 KQ562558 KXJ75630.1 NWSH01000098 PCG79585.1 ODYU01012071 SOQ58176.1 JXUM01021492 KQ560602 KXJ81676.1 KZ149975 PZC75948.1 JRES01001303 KNC23770.1 PCG72679.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JVL1
A0A2W1BUU8
A0A2H1V250
A0A194Q1T6
A0A212EY43
A0A2H1WI98
+ More
A0A2A4JL38 A0A0L7K4N0 A0A194Q2D2 H9JVM2 A0A0N1IG89 A0A2A4JAH3 A0A194Q800 A0A194Q1Q2 A0A2A4IYX6 A0A194Q1T1 A0A2H1WJU4 A0A2W1BVQ5 A0A0L7L435 A0A212FGQ8 A0A2H1V0R1 A0A212F5C2 A0A2H1VUC9 A0A2A4J999 A0A0N0PCK2 A0A2W1BGE8 H9JV72 A0A2H1W801 S4Q006 A0A0N1I9N3 A0A2A4JT62 A0A2H1WUV1 A0A2W1BZI9 A0A2H1VEB4 A0A0L7LHZ8 A0A2H1VUK7 A0A2H1WU82 A0A2H1VVE4 A0A0N0PCN2 A0A2W1BWY0 H9JVL3 A0A2H1WGI5 A0A2A4JX40 A0A194Q1R5 A0A212EQC0 A0A0N1IPG1 H9JVR6 A0A2H1W7K7 A0A1Q3FNL7 A0A1Q3FNK2 A0A1I8P9Y1 B0W415 A0A2A4JKW1 A0A2A4JLN1 A0A1Q3FM89 A0A1Q3FLR3 A0A034VU45 T1GER7 A0A034VUT9 A0A2H1X0I9 A0A0K8UIB9 A0A1I8P1U8 A0A1I8P1R5 A0A1L8E785 W8B9U3 A0A1L8E7E8 A0A1L8E7J3 A0A0A1XE60 W8B5D0 A0A1I8MVX6 A0A1L8EFQ6 A0A1I8MVY3 A0A1L8EFY3 T1PCW5 A0A1L8EFU2 A0A212EZN8 A0A194PJI3 B0WKE4 A0A2A4JLT5 A0A0A1XC67 A0A1A9X005 A0A212EYD8 U5ETH9 A0A1I8MZK1 A0A182GKA8 A0A2A4K5Q7 A0A2H1WYY1 A0A182GYQ4 A0A2W1BLZ7 A0A0L0BUM3 A0A2A4JLI7 A0A1A9WGN7
A0A2A4JL38 A0A0L7K4N0 A0A194Q2D2 H9JVM2 A0A0N1IG89 A0A2A4JAH3 A0A194Q800 A0A194Q1Q2 A0A2A4IYX6 A0A194Q1T1 A0A2H1WJU4 A0A2W1BVQ5 A0A0L7L435 A0A212FGQ8 A0A2H1V0R1 A0A212F5C2 A0A2H1VUC9 A0A2A4J999 A0A0N0PCK2 A0A2W1BGE8 H9JV72 A0A2H1W801 S4Q006 A0A0N1I9N3 A0A2A4JT62 A0A2H1WUV1 A0A2W1BZI9 A0A2H1VEB4 A0A0L7LHZ8 A0A2H1VUK7 A0A2H1WU82 A0A2H1VVE4 A0A0N0PCN2 A0A2W1BWY0 H9JVL3 A0A2H1WGI5 A0A2A4JX40 A0A194Q1R5 A0A212EQC0 A0A0N1IPG1 H9JVR6 A0A2H1W7K7 A0A1Q3FNL7 A0A1Q3FNK2 A0A1I8P9Y1 B0W415 A0A2A4JKW1 A0A2A4JLN1 A0A1Q3FM89 A0A1Q3FLR3 A0A034VU45 T1GER7 A0A034VUT9 A0A2H1X0I9 A0A0K8UIB9 A0A1I8P1U8 A0A1I8P1R5 A0A1L8E785 W8B9U3 A0A1L8E7E8 A0A1L8E7J3 A0A0A1XE60 W8B5D0 A0A1I8MVX6 A0A1L8EFQ6 A0A1I8MVY3 A0A1L8EFY3 T1PCW5 A0A1L8EFU2 A0A212EZN8 A0A194PJI3 B0WKE4 A0A2A4JLT5 A0A0A1XC67 A0A1A9X005 A0A212EYD8 U5ETH9 A0A1I8MZK1 A0A182GKA8 A0A2A4K5Q7 A0A2H1WYY1 A0A182GYQ4 A0A2W1BLZ7 A0A0L0BUM3 A0A2A4JLI7 A0A1A9WGN7
Ontologies
GO
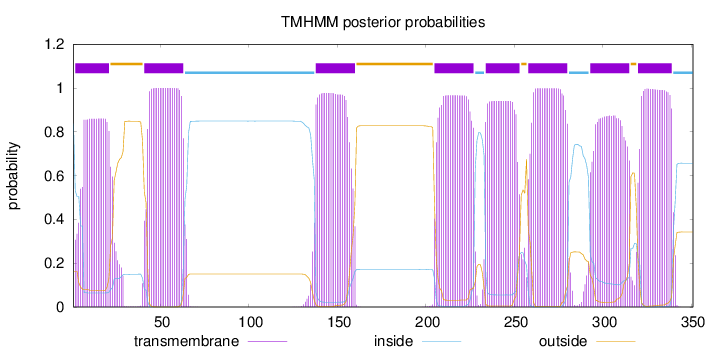
Topology
Length:
351
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
163.17686
Exp number, first 60 AAs:
36.19623
Total prob of N-in:
0.83800
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 21
outside
22 - 40
TMhelix
41 - 63
inside
64 - 137
TMhelix
138 - 160
outside
161 - 204
TMhelix
205 - 227
inside
228 - 233
TMhelix
234 - 253
outside
254 - 257
TMhelix
258 - 280
inside
281 - 292
TMhelix
293 - 315
outside
316 - 319
TMhelix
320 - 339
inside
340 - 351
Population Genetic Test Statistics
Pi
189.06301
Theta
175.884641
Tajima's D
0.736551
CLR
1.539269
CSRT
0.585320733963302
Interpretation
Uncertain
