Gene
KWMTBOMO03197
Pre Gene Modal
BGIBMGA013573
Annotation
PREDICTED:_cadherin-87A_[Bombyx_mori]
Full name
Cadherin-87A
Location in the cell
Cytoplasmic Reliability : 1.275 Nuclear Reliability : 1.539
Sequence
CDS
ATGGAAACAAGATGGAGAACGTTCGTGATATTATCGTTCTGCTTGGGTTCAAGCTTAGCAAACATGTTACCGGTGTTCACGCAAGACATGAACAACTTACCGCTATCGGAGAGTACTCCGGTTGGCACAGTGGTCTACACACTGCAGGGAGTTCACCCTCAGGGGTTACCGATCAAATACAGCATGGTCGGCTCTGACAATTTCGTTGTAGATCCTGAGTCCGGCGATGTCACGCTGGACAAGCCTTTGGACAGGGAGGTTGACGATACGCTGAAGTTCCTCGTTTCCATTGAAGCTGCCGACCCTCAGGGTCAGCAGAAGCTGATACAGAAACAGCCCGTCACAGTCATAGTGCTAGATGAAAATGATAACCCGCCTACATTCAAAAACAACCCTTACGAAGTAGACGTGCCAGAAGACGAAGCAATAGGAACGACCATCCTCAACAACATACTGGTGGAAGATAAAGATTTTGTCGGCGATAATCTTGAAGTTGGATGTTCGGCCAATGACCAGTGGCCTGAAGCTTGCGATATCTTTGACGTAGTGGCTTTAAACTCGACTTCGCACCACTACCACGGAGCTCTGGTCCTGAAGAGGCCCCTCAATTACAACGAAAGGCAGTTCTATCAGTTTCAATTGTCTGCCACAGACGGCATTCAGAACTCCTCCGGACAAGTAGAAGTGAAGGTGATAGACGTCCAGAATTCGAGTCCCATGTTCTACGGAGCTCTCAGCGGAGCGCTGTCGGAGGACGCGCCGGTCGGCACCCTGGTGCTGACGGTGCAGGCCAGGGACGCTGACAAGGCTCAACCCAGGCCTGTCGTCTTGGAGCTGCTCACTAATCCTCTGGATTTCTTTCTGTTGGACAAAAACACTGGAGAACTACGGACAGCTAAGCCGCTAGATAGAGAAGCTTTAGACGATCCGTCCTCGCCACTAAATTTAACCGTCAGGGCAACCGAAATAGTGAATGGCTCACCGTTGATATCGGATTTGACGTCGACCGTCGCGACGGTGACCATCACCATTAAGGACGTGAACGACGAACCGCCCCGCTTCAACCGAAGGGAGTACGCCGTTCAGATACCGGAGAGTCTACCGGTCGGGACGCCGCTGCCCAACTTAGAGATGGTCGTCACCGACACTGATTTGGGTCTCAATTCGGTGTTCGGTTTGCGTTTACTGAATGAGTTCGAAGCGTTCGTCGTGGAGCCCCCGTCGGCAACCGGCAGCGCCTCCGTCACCATACGACTCAACTCCAGCTTGGACTACGAGGACCCGAACCAGAGGAAGTTCATTTTGGAGGTGATAGCAGAAGAGCTGCACACGACCCCGAAGCTGTCGTCACGGGCCAGCGTGACGGTGACAGTGACAGACGTCAATGACAATGCGCCGCAATTCGCGGAGGAGCCCGTGGTGCGGTCCTTATCGGAGACTTCGCCCCCCGGCACCCGCCTCATTGCCTCGCGTGCCACTGACCGGGACACGGGAAGGTTCGGTACTGAAGGTATTGTGTATGAGCTCTCCGGTAACGGGGCGGAGCTGTTTGACGTCGACAAGAGATCTGGTGTGATCAGCGTCGCGGAATGCCCTACTCCTGGACAACCCCCGTGCCTGGACTACGAGAGCAGAAAGGACTATTTCCTGCAATATAAGGCGACTGATGATTACGGCAACGGTCAAATGACGGTGGTCTCGCTTCAGATATCATTGATTGATGCGAACGACAATGCCCCAGTATTTGTGACGCCGGTATACAAGGCGTCCATTGACGAAGACGCTGTCAAATTCGAACCAGAACTACAGGTTCTAGCCCGCGATGTCGACGTGTCATCAGACATACGCTACTCTATCGTGGGTCCGGCGACGCATCCGTTCTGGATCGATCCAGTCACCGGGAAGATCACAGTCAACCCTGAGTCCGGGAGAGTGGCAGCGGTGGATGACTCCAACAAGATAATACTCACTGTATTGACGGAGGAGTATGAAATTTTCCACACTTATAGAGAATATAGAGAAGAAGTGCACAATGATAATATTTTTTTAAAATAA
Protein
METRWRTFVILSFCLGSSLANMLPVFTQDMNNLPLSESTPVGTVVYTLQGVHPQGLPIKYSMVGSDNFVVDPESGDVTLDKPLDREVDDTLKFLVSIEAADPQGQQKLIQKQPVTVIVLDENDNPPTFKNNPYEVDVPEDEAIGTTILNNILVEDKDFVGDNLEVGCSANDQWPEACDIFDVVALNSTSHHYHGALVLKRPLNYNERQFYQFQLSATDGIQNSSGQVEVKVIDVQNSSPMFYGALSGALSEDAPVGTLVLTVQARDADKAQPRPVVLELLTNPLDFFLLDKNTGELRTAKPLDREALDDPSSPLNLTVRATEIVNGSPLISDLTSTVATVTITIKDVNDEPPRFNRREYAVQIPESLPVGTPLPNLEMVVTDTDLGLNSVFGLRLLNEFEAFVVEPPSATGSASVTIRLNSSLDYEDPNQRKFILEVIAEELHTTPKLSSRASVTVTVTDVNDNAPQFAEEPVVRSLSETSPPGTRLIASRATDRDTGRFGTEGIVYELSGNGAELFDVDKRSGVISVAECPTPGQPPCLDYESRKDYFLQYKATDDYGNGQMTVVSLQISLIDANDNAPVFVTPVYKASIDEDAVKFEPELQVLARDVDVSSDIRYSIVGPATHPFWIDPVTGKITVNPESGRVAAVDDSNKIILTVLTEEYEIFHTYREYREEVHNDNIFLK
Summary
Description
Cadherins are calcium-dependent cell adhesion proteins. They preferentially interact with themselves in a homophilic manner in connecting cells (By similarity).
Keywords
Calcium
Cell adhesion
Cell membrane
Complete proteome
Glycoprotein
Membrane
Metal-binding
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Cadherin-87A
Uniprot
H9JVL0
A0A2H1VJV4
A0A194Q1R2
A0A212EXY9
A0A0N0PBZ6
A0A1B6L3B2
+ More
A0A1B6G2W1 A0A1B6BWB3 A0A3L8DTJ8 A0A2L1IQ80 A0A0N0U6S0 A0A158P1K1 A0A067R2V0 A0A0C9RKC3 A0A2A3E7T2 A0A087ZWQ8 A0A151I809 A0A146KXI2 A0A0A9X5Z0 A0A151XGS7 A0A232FF41 T1I8I2 D6WC50 A0A0T6B338 E0W2T6 A0A195DUL5 A0A0J7NR78 E2AGF4 A0A151I3A9 A0A2R7VSP5 A0A1B6JL50 A0A2J7R340 A0A2P6L2Z9 A0A087UQR6 A0A2P8Z823 U4UPW6 A0A1Y1MCH0 A0A0L0CE80 E2BG92 A0A1I8MKU2 A0A1I8MKX7 A0A1I8MKU6 A0A1I8MKU3 A0A147BFY1 A0A0K8U123 W8BEQ1 A0A2S2Q7P4 A0A1I8NS02 A0A1I8NRZ8 A0A2H8TK70 A0A0L7RI16 A0A336M3D8 X1WQ77 A0A2S2NVT6 A0A1S4GIJ5 A0A0K8VZZ4 A0A0K8UJS3 A0A1Y9GKC3 A0A2C9H8D3 B3MTU1 A0A0P8XDJ2 A0A182KXI7 Q7QAQ8 A0A1W4W1A8 B4JG13 B5DVW9 Q9VGG5 A0A0Q9WWP4 B4LXZ1 A0A3B0JN51 B4K9Z6 B4QTJ8 A0A0M4EQ25 A0A1J1HU01 B4HHK1 B0W917 B4PTE6 B3P4D8 A0A154PK18 A0A1A9VG57 A0A1B0FHA7 A0A1A9YHW0 N6SYU5 A0A2W1BD34 A0A1A9ZYV1 A0A1A9WEW2 A0A195EXJ7 A0A1B0B9U7 A0A0K2TYX2 A0A1D2MQG8
A0A1B6G2W1 A0A1B6BWB3 A0A3L8DTJ8 A0A2L1IQ80 A0A0N0U6S0 A0A158P1K1 A0A067R2V0 A0A0C9RKC3 A0A2A3E7T2 A0A087ZWQ8 A0A151I809 A0A146KXI2 A0A0A9X5Z0 A0A151XGS7 A0A232FF41 T1I8I2 D6WC50 A0A0T6B338 E0W2T6 A0A195DUL5 A0A0J7NR78 E2AGF4 A0A151I3A9 A0A2R7VSP5 A0A1B6JL50 A0A2J7R340 A0A2P6L2Z9 A0A087UQR6 A0A2P8Z823 U4UPW6 A0A1Y1MCH0 A0A0L0CE80 E2BG92 A0A1I8MKU2 A0A1I8MKX7 A0A1I8MKU6 A0A1I8MKU3 A0A147BFY1 A0A0K8U123 W8BEQ1 A0A2S2Q7P4 A0A1I8NS02 A0A1I8NRZ8 A0A2H8TK70 A0A0L7RI16 A0A336M3D8 X1WQ77 A0A2S2NVT6 A0A1S4GIJ5 A0A0K8VZZ4 A0A0K8UJS3 A0A1Y9GKC3 A0A2C9H8D3 B3MTU1 A0A0P8XDJ2 A0A182KXI7 Q7QAQ8 A0A1W4W1A8 B4JG13 B5DVW9 Q9VGG5 A0A0Q9WWP4 B4LXZ1 A0A3B0JN51 B4K9Z6 B4QTJ8 A0A0M4EQ25 A0A1J1HU01 B4HHK1 B0W917 B4PTE6 B3P4D8 A0A154PK18 A0A1A9VG57 A0A1B0FHA7 A0A1A9YHW0 N6SYU5 A0A2W1BD34 A0A1A9ZYV1 A0A1A9WEW2 A0A195EXJ7 A0A1B0B9U7 A0A0K2TYX2 A0A1D2MQG8
Pubmed
19121390
26354079
22118469
30249741
29415259
21347285
+ More
24845553 26823975 25401762 28648823 18362917 19820115 20566863 20798317 29403074 23537049 28004739 26108605 25315136 29652888 24495485 12364791 17994087 18057021 20966253 15632085 10731132 12537572 12537569 17893096 17550304 28756777 27289101
24845553 26823975 25401762 28648823 18362917 19820115 20566863 20798317 29403074 23537049 28004739 26108605 25315136 29652888 24495485 12364791 17994087 18057021 20966253 15632085 10731132 12537572 12537569 17893096 17550304 28756777 27289101
EMBL
BABH01032472
BABH01032473
BABH01032474
BABH01032475
BABH01032476
BABH01032477
+ More
BABH01032478 ODYU01002963 SOQ41129.1 KQ459579 KPI99517.1 AGBW02011640 OWR46355.1 KQ460757 KPJ12580.1 GEBQ01021841 JAT18136.1 GECZ01012991 JAS56778.1 GEDC01031725 JAS05573.1 QOIP01000004 RLU23483.1 MF741659 AVD96942.1 KQ435729 KOX77608.1 ADTU01006385 ADTU01006386 ADTU01006387 ADTU01006388 KK852798 KDR16370.1 GBYB01013722 JAG83489.1 KZ288354 PBC27276.1 KQ978415 KYM94087.1 GDHC01018882 JAP99746.1 GBHO01029376 JAG14228.1 KQ982138 KYQ59612.1 NNAY01000333 OXU29140.1 ACPB03019929 KQ971316 EEZ99177.2 LJIG01016077 KRT81684.1 DS235879 EEB19942.1 KQ980390 KYN16244.1 LBMM01002334 KMQ94965.1 GL439310 EFN67487.1 KQ976512 KYM82427.1 KK854063 PTY10524.1 GECU01007748 JAS99958.1 NEVH01007822 PNF35261.1 MWRG01002194 PRD32960.1 KK121082 KFM79705.1 PYGN01000154 PSN52632.1 KB632314 ERL92186.1 GEZM01034916 JAV83532.1 JRES01000630 KNC29789.1 GL448116 EFN85300.1 GEGO01005726 JAR89678.1 GDHF01031875 JAI20439.1 GAMC01014864 JAB91691.1 GGMS01004553 MBY73756.1 GFXV01002247 MBW14052.1 KQ414588 KOC70438.1 UFQS01000496 UFQT01000496 SSX04398.1 SSX24762.1 ABLF02020103 GGMR01008489 MBY21108.1 AAAB01008888 GDHF01026882 GDHF01007905 JAI25432.1 JAI44409.1 GDHF01025714 JAI26600.1 APCN01005528 CH902623 EDV30222.1 KPU72743.1 KPU72744.1 KPU72742.1 EAA08909.4 CH916369 EDV92552.1 CM000070 EDY68473.1 AE014297 AY128505 CH964272 KRG00181.1 CH940650 EDW66857.2 KRF82945.1 OUUW01000008 SPP83644.1 CH933806 EDW15644.2 KRG01652.1 CM000364 EDX13303.1 CP012526 ALC46335.1 CVRI01000021 CRK91477.1 CH480815 EDW42540.1 DS231861 EDS39455.1 CM000160 EDW97645.1 KRK03874.1 KRK03875.1 CH954181 EDV49453.1 KQ434942 KZC12192.1 CCAG010019049 APGK01051728 APGK01051729 APGK01051730 APGK01051731 KB741202 ENN72944.1 KZ150255 PZC71795.1 KQ981928 KYN32871.1 JXJN01010593 HACA01013210 CDW30571.1 LJIJ01000716 ODM95064.1
BABH01032478 ODYU01002963 SOQ41129.1 KQ459579 KPI99517.1 AGBW02011640 OWR46355.1 KQ460757 KPJ12580.1 GEBQ01021841 JAT18136.1 GECZ01012991 JAS56778.1 GEDC01031725 JAS05573.1 QOIP01000004 RLU23483.1 MF741659 AVD96942.1 KQ435729 KOX77608.1 ADTU01006385 ADTU01006386 ADTU01006387 ADTU01006388 KK852798 KDR16370.1 GBYB01013722 JAG83489.1 KZ288354 PBC27276.1 KQ978415 KYM94087.1 GDHC01018882 JAP99746.1 GBHO01029376 JAG14228.1 KQ982138 KYQ59612.1 NNAY01000333 OXU29140.1 ACPB03019929 KQ971316 EEZ99177.2 LJIG01016077 KRT81684.1 DS235879 EEB19942.1 KQ980390 KYN16244.1 LBMM01002334 KMQ94965.1 GL439310 EFN67487.1 KQ976512 KYM82427.1 KK854063 PTY10524.1 GECU01007748 JAS99958.1 NEVH01007822 PNF35261.1 MWRG01002194 PRD32960.1 KK121082 KFM79705.1 PYGN01000154 PSN52632.1 KB632314 ERL92186.1 GEZM01034916 JAV83532.1 JRES01000630 KNC29789.1 GL448116 EFN85300.1 GEGO01005726 JAR89678.1 GDHF01031875 JAI20439.1 GAMC01014864 JAB91691.1 GGMS01004553 MBY73756.1 GFXV01002247 MBW14052.1 KQ414588 KOC70438.1 UFQS01000496 UFQT01000496 SSX04398.1 SSX24762.1 ABLF02020103 GGMR01008489 MBY21108.1 AAAB01008888 GDHF01026882 GDHF01007905 JAI25432.1 JAI44409.1 GDHF01025714 JAI26600.1 APCN01005528 CH902623 EDV30222.1 KPU72743.1 KPU72744.1 KPU72742.1 EAA08909.4 CH916369 EDV92552.1 CM000070 EDY68473.1 AE014297 AY128505 CH964272 KRG00181.1 CH940650 EDW66857.2 KRF82945.1 OUUW01000008 SPP83644.1 CH933806 EDW15644.2 KRG01652.1 CM000364 EDX13303.1 CP012526 ALC46335.1 CVRI01000021 CRK91477.1 CH480815 EDW42540.1 DS231861 EDS39455.1 CM000160 EDW97645.1 KRK03874.1 KRK03875.1 CH954181 EDV49453.1 KQ434942 KZC12192.1 CCAG010019049 APGK01051728 APGK01051729 APGK01051730 APGK01051731 KB741202 ENN72944.1 KZ150255 PZC71795.1 KQ981928 KYN32871.1 JXJN01010593 HACA01013210 CDW30571.1 LJIJ01000716 ODM95064.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000279307
UP000053105
+ More
UP000005205 UP000027135 UP000242457 UP000005203 UP000078542 UP000075809 UP000215335 UP000015103 UP000007266 UP000009046 UP000078492 UP000036403 UP000000311 UP000078540 UP000235965 UP000054359 UP000245037 UP000030742 UP000037069 UP000008237 UP000095301 UP000095300 UP000053825 UP000007819 UP000075840 UP000076407 UP000007801 UP000075882 UP000007062 UP000192221 UP000001070 UP000001819 UP000000803 UP000007798 UP000008792 UP000268350 UP000009192 UP000000304 UP000092553 UP000183832 UP000001292 UP000002320 UP000002282 UP000008711 UP000076502 UP000078200 UP000092444 UP000092443 UP000019118 UP000092445 UP000091820 UP000078541 UP000092460 UP000094527
UP000005205 UP000027135 UP000242457 UP000005203 UP000078542 UP000075809 UP000215335 UP000015103 UP000007266 UP000009046 UP000078492 UP000036403 UP000000311 UP000078540 UP000235965 UP000054359 UP000245037 UP000030742 UP000037069 UP000008237 UP000095301 UP000095300 UP000053825 UP000007819 UP000075840 UP000076407 UP000007801 UP000075882 UP000007062 UP000192221 UP000001070 UP000001819 UP000000803 UP000007798 UP000008792 UP000268350 UP000009192 UP000000304 UP000092553 UP000183832 UP000001292 UP000002320 UP000002282 UP000008711 UP000076502 UP000078200 UP000092444 UP000092443 UP000019118 UP000092445 UP000091820 UP000078541 UP000092460 UP000094527
Pfam
PF00028 Cadherin
SUPFAM
SSF49313
SSF49313
ProteinModelPortal
H9JVL0
A0A2H1VJV4
A0A194Q1R2
A0A212EXY9
A0A0N0PBZ6
A0A1B6L3B2
+ More
A0A1B6G2W1 A0A1B6BWB3 A0A3L8DTJ8 A0A2L1IQ80 A0A0N0U6S0 A0A158P1K1 A0A067R2V0 A0A0C9RKC3 A0A2A3E7T2 A0A087ZWQ8 A0A151I809 A0A146KXI2 A0A0A9X5Z0 A0A151XGS7 A0A232FF41 T1I8I2 D6WC50 A0A0T6B338 E0W2T6 A0A195DUL5 A0A0J7NR78 E2AGF4 A0A151I3A9 A0A2R7VSP5 A0A1B6JL50 A0A2J7R340 A0A2P6L2Z9 A0A087UQR6 A0A2P8Z823 U4UPW6 A0A1Y1MCH0 A0A0L0CE80 E2BG92 A0A1I8MKU2 A0A1I8MKX7 A0A1I8MKU6 A0A1I8MKU3 A0A147BFY1 A0A0K8U123 W8BEQ1 A0A2S2Q7P4 A0A1I8NS02 A0A1I8NRZ8 A0A2H8TK70 A0A0L7RI16 A0A336M3D8 X1WQ77 A0A2S2NVT6 A0A1S4GIJ5 A0A0K8VZZ4 A0A0K8UJS3 A0A1Y9GKC3 A0A2C9H8D3 B3MTU1 A0A0P8XDJ2 A0A182KXI7 Q7QAQ8 A0A1W4W1A8 B4JG13 B5DVW9 Q9VGG5 A0A0Q9WWP4 B4LXZ1 A0A3B0JN51 B4K9Z6 B4QTJ8 A0A0M4EQ25 A0A1J1HU01 B4HHK1 B0W917 B4PTE6 B3P4D8 A0A154PK18 A0A1A9VG57 A0A1B0FHA7 A0A1A9YHW0 N6SYU5 A0A2W1BD34 A0A1A9ZYV1 A0A1A9WEW2 A0A195EXJ7 A0A1B0B9U7 A0A0K2TYX2 A0A1D2MQG8
A0A1B6G2W1 A0A1B6BWB3 A0A3L8DTJ8 A0A2L1IQ80 A0A0N0U6S0 A0A158P1K1 A0A067R2V0 A0A0C9RKC3 A0A2A3E7T2 A0A087ZWQ8 A0A151I809 A0A146KXI2 A0A0A9X5Z0 A0A151XGS7 A0A232FF41 T1I8I2 D6WC50 A0A0T6B338 E0W2T6 A0A195DUL5 A0A0J7NR78 E2AGF4 A0A151I3A9 A0A2R7VSP5 A0A1B6JL50 A0A2J7R340 A0A2P6L2Z9 A0A087UQR6 A0A2P8Z823 U4UPW6 A0A1Y1MCH0 A0A0L0CE80 E2BG92 A0A1I8MKU2 A0A1I8MKX7 A0A1I8MKU6 A0A1I8MKU3 A0A147BFY1 A0A0K8U123 W8BEQ1 A0A2S2Q7P4 A0A1I8NS02 A0A1I8NRZ8 A0A2H8TK70 A0A0L7RI16 A0A336M3D8 X1WQ77 A0A2S2NVT6 A0A1S4GIJ5 A0A0K8VZZ4 A0A0K8UJS3 A0A1Y9GKC3 A0A2C9H8D3 B3MTU1 A0A0P8XDJ2 A0A182KXI7 Q7QAQ8 A0A1W4W1A8 B4JG13 B5DVW9 Q9VGG5 A0A0Q9WWP4 B4LXZ1 A0A3B0JN51 B4K9Z6 B4QTJ8 A0A0M4EQ25 A0A1J1HU01 B4HHK1 B0W917 B4PTE6 B3P4D8 A0A154PK18 A0A1A9VG57 A0A1B0FHA7 A0A1A9YHW0 N6SYU5 A0A2W1BD34 A0A1A9ZYV1 A0A1A9WEW2 A0A195EXJ7 A0A1B0B9U7 A0A0K2TYX2 A0A1D2MQG8
PDB
5W4T
E-value=1.0776e-23,
Score=275
Ontologies
GO
Topology
Subcellular location
Cell membrane
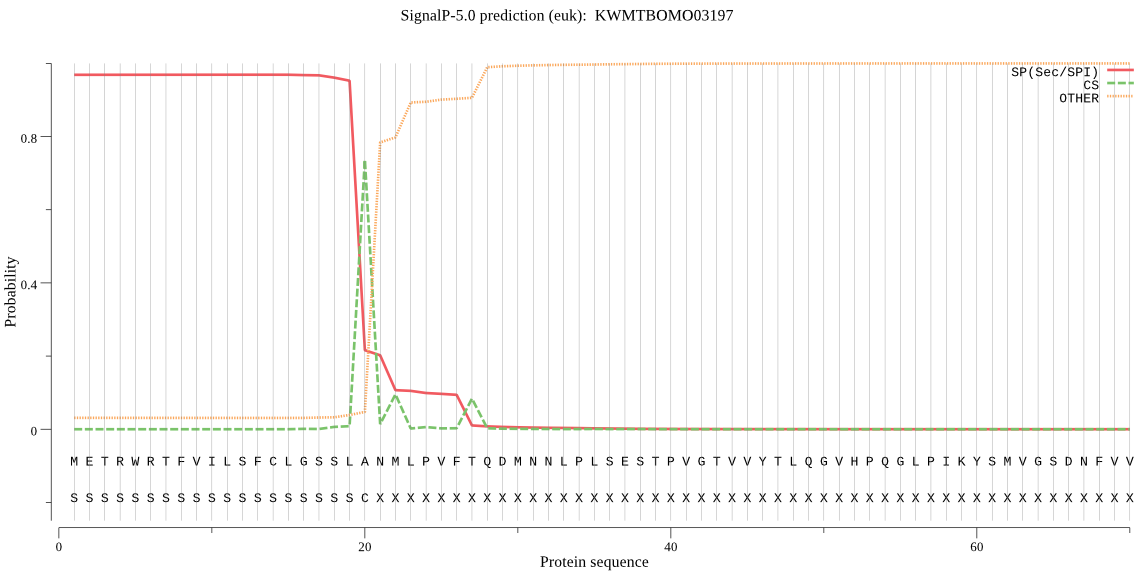
SignalP
Position: 1 - 20,
Likelihood: 0.968525
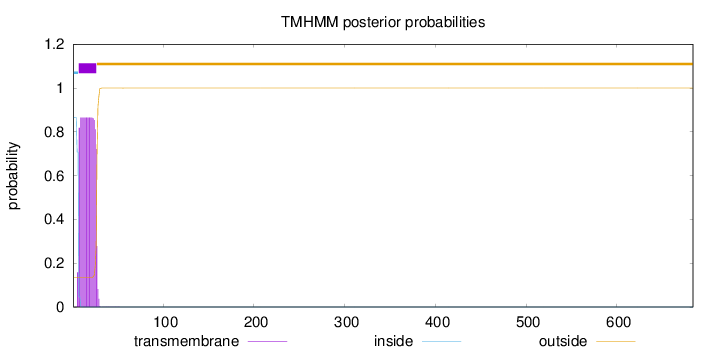
Length:
684
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.71915
Exp number, first 60 AAs:
17.71892
Total prob of N-in:
0.86535
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 684
Population Genetic Test Statistics
Pi
327.845572
Theta
183.852188
Tajima's D
2.430907
CLR
0.418029
CSRT
0.937453127343633
Interpretation
Uncertain
