Gene
KWMTBOMO03194
Annotation
PREDICTED:_uncharacterized_protein_LOC101735858_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.811 PlasmaMembrane Reliability : 1.889
Sequence
CDS
ATGGGAGACTTCAAAACCGACCTCCTTGTCCCTTTGTCCACTCGTTTACGAAAACTCCTTGACCTTGTTGAGTCAGTTAGTTTACACATCCTCCCTTTACTAGCCACTCACCACAACATTGAAGGAGAGGATACCTGGCTTGACCTAATACTCACTTCCAATCCCGATCTTGTTCATTCTCACGGTCAGTTTCCTGCTCCTGGTTTCTCCTACCATGACCTCATTTACTTATCCTACGTCCTTAAACCTCCCAAATCCAAGCCTAAAGTTTTACGCGTGCGTTGTTTTGGTCGCATGGATGGACGCAGGTTGTGTGAGGATGCTGCTAAAATTAGTTGGGATGAACTGACGGCAGCTACATCTATTGACGACAAGGTGACTATATTTAACGCTCAACTCCAGGCACTTTATGACACTCATGCTCCCGTCAGGAAATTCAAGTTGAAACGTCCTCCAGCACCTTGGATGACGAGTGAGGTTCGGATGGCGATGAAGCGTAGGGATCGGGCCTTGGCAAAATTCAGGAGACGCCGCACGGAGGAAAACTGGACATCATTTAAGGTTGCGACGAATCGGTGTAATCAGATGGTACGGAACGCTAAGCGCCGCCATATTCTCGAAAATATAACCTCCGCTTCTTCAGCGAATATCTGGAAGTTCTTTAAGACTCTGGGTACTGGTAGGGAGAAGAATAAAGACTTCCAGGGGACCATTGCCTTAGATGACCTTAACAGGCATTTCAGCGCATCTGCCGGCATTGATCATCAAGTCAAAAGTCTCACTATTGAATTTATAGCTGGGTTACCTAAACCAGACATTGACCCCTTCCAATTTTCTCCAATGACTTCAGATGATATTAAAAAGATTATCCTATCCATTAGATCTAATGCTGTTGGTTATGACAAAATCACCCATCGCATGATTACCACAGCTTTAGACCATCTTATTCTCACCATAACCCACATCATTAATTTCTCCTTTAGTACCAACATGTTTCCCTCCCAATGGCGTAAGGCTTATGTCATTCCTCTTCCTAAAATACCGAATCCTACCCTTGTTACTCATTTCCGACCTATATCCATACTTCCTTTCCTATCTAAGGTTTTAGAAGCTTGTGCGCACAAACAACTGTCCAATTTTATTCATTCACAACATTTGTTGAGTACATTGCAGTCCGGTTTCAGACCTTGCCACAGCACTACTACCGCACTCCTTAAAGTGATTGGCGATATTAGAGCAGGTATGGAGGACACAAAGGTTACGGTATTGGTTCTGATAGACTTCTCAAACGCTTTTAATAACGTCAATCACGACATCCTCTTATCCATTCTCTCCCATCTTAGGATGTCATCTGGAGCATTGGATTGGTTCTCGTCTTATCTTCGAGGTCGTCAGCAGTCGATACACATCAACGAGTTTTCGTCTAGTTGGTGTGATCTGCACTCTGGCGTTCCTCAAGGCGGCATTCTTTCTCCTCTACTATTTTCAATTTTTATTAATCTCATTACCGATAATCTTCAATGTGCGTACCATCTATATGCCGATGATTTGCAGCTTTATGCTCAGGCTGGAGTCGATAATGTGTCTTCGGCAGTTGGTAAAATAAATAACGACCTGTCCTACATTAAATTCTGGTCTGACAGTTTCGGCTTAAGCGTGAATCCCGCCAAATGTCAGGCTATTATTGTTGGAAGTCAACGCTTGTTGTATCGGTTGGATATGGCATATGTCCCATCGGTTATATATGATGGCTCAACCATCAACTGGTGTTCAGTTGTAAAGGACCTAGGCCTACTTATTGACTCCAGCCTGAGCTGGCGAGGTCAGGCCCAGGTCATCTCCGTGTGTCAGCGAGTTACACTTACACTCCGTATGCTTTACCGCTGA
Protein
MGDFKTDLLVPLSTRLRKLLDLVESVSLHILPLLATHHNIEGEDTWLDLILTSNPDLVHSHGQFPAPGFSYHDLIYLSYVLKPPKSKPKVLRVRCFGRMDGRRLCEDAAKISWDELTAATSIDDKVTIFNAQLQALYDTHAPVRKFKLKRPPAPWMTSEVRMAMKRRDRALAKFRRRRTEENWTSFKVATNRCNQMVRNAKRRHILENITSASSANIWKFFKTLGTGREKNKDFQGTIALDDLNRHFSASAGIDHQVKSLTIEFIAGLPKPDIDPFQFSPMTSDDIKKIILSIRSNAVGYDKITHRMITTALDHLILTITHIINFSFSTNMFPSQWRKAYVIPLPKIPNPTLVTHFRPISILPFLSKVLEACAHKQLSNFIHSQHLLSTLQSGFRPCHSTTTALLKVIGDIRAGMEDTKVTVLVLIDFSNAFNNVNHDILLSILSHLRMSSGALDWFSSYLRGRQQSIHINEFSSSWCDLHSGVPQGGILSPLLFSIFINLITDNLQCAYHLYADDLQLYAQAGVDNVSSAVGKINNDLSYIKFWSDSFGLSVNPAKCQAIIVGSQRLLYRLDMAYVPSVIYDGSTINWCSVVKDLGLLIDSSLSWRGQAQVISVCQRVTLTLRMLYR
Summary
Uniprot
A0A2A4JRM8
A0A1B6L6Y3
A0A2A4JPY6
A0A0J7K589
A0A0J7K195
A0A0A9WX05
+ More
A0A1B6LTE5 A0A0A9YIX2 A0A0J7N335 A0A3C1S1N3 A0A1B6KKS7 A0A023EXE1 A0A3C1S2P1 A0A354GJG9 A0A1W7R6F6 U5EQQ2 A0A0J7KR96 A0A3C1RZS6 A0A354GES4 A0A3C1RY27 A0A354GIY0 A0A1S3DPM6 U5EUG4 U5ENG8 W8AIM4 W8B6W6 A0A3L8DSB3 U5EQV9 U5EQB9 A0A3C1S360 U5EQA6 A0A1B6KD49 U5EEC7 U5EN99 A0A354GG23 A0A1Y1L0L4 U5ETR7 A0A2B4RHP5 A0A1W7R648 A0A1W7R6J3 U5EEL4 A0A3C1RYE8 A0A1W7R6K8 U5ER05 A0A1W7R6E4 A0A1Y1M3U5 T1DE06 A0A354GHC6 A0A1Y1KZ78 A0A1Y1NB24 K7JTK2 A0A1B6IH82 U5EQT8 U5EQE2 U5ETW8 U5EVC6 K7JAP1 A0A1Y1N9S9 A0A1Y1NKD6 A0A2A4JF59 A0A1Y1LSZ2 A0A1L8DJB0 A0A2W1BK42 A0A1Y1LVZ3 A0A3B3HNZ2 A0A2A4KAV4 U5EF12 A0A0A9YBT8 A0A3B3BNA3 K7JBA8 Q4F8Q2 U5EQJ2 A0A0J7KSZ7 A0A2B4SN01 A0A2A4JM54 A0A3B3HIF3 A0A2B4RRX2
A0A1B6LTE5 A0A0A9YIX2 A0A0J7N335 A0A3C1S1N3 A0A1B6KKS7 A0A023EXE1 A0A3C1S2P1 A0A354GJG9 A0A1W7R6F6 U5EQQ2 A0A0J7KR96 A0A3C1RZS6 A0A354GES4 A0A3C1RY27 A0A354GIY0 A0A1S3DPM6 U5EUG4 U5ENG8 W8AIM4 W8B6W6 A0A3L8DSB3 U5EQV9 U5EQB9 A0A3C1S360 U5EQA6 A0A1B6KD49 U5EEC7 U5EN99 A0A354GG23 A0A1Y1L0L4 U5ETR7 A0A2B4RHP5 A0A1W7R648 A0A1W7R6J3 U5EEL4 A0A3C1RYE8 A0A1W7R6K8 U5ER05 A0A1W7R6E4 A0A1Y1M3U5 T1DE06 A0A354GHC6 A0A1Y1KZ78 A0A1Y1NB24 K7JTK2 A0A1B6IH82 U5EQT8 U5EQE2 U5ETW8 U5EVC6 K7JAP1 A0A1Y1N9S9 A0A1Y1NKD6 A0A2A4JF59 A0A1Y1LSZ2 A0A1L8DJB0 A0A2W1BK42 A0A1Y1LVZ3 A0A3B3HNZ2 A0A2A4KAV4 U5EF12 A0A0A9YBT8 A0A3B3BNA3 K7JBA8 Q4F8Q2 U5EQJ2 A0A0J7KSZ7 A0A2B4SN01 A0A2A4JM54 A0A3B3HIF3 A0A2B4RRX2
Pubmed
EMBL
NWSH01000817
PCG74070.1
GEBQ01020572
JAT19405.1
NWSH01000893
PCG73654.1
+ More
LBMM01014048 KMQ85356.1 LBMM01017805 KMQ83966.1 GBHO01031326 GBHO01031325 JAG12278.1 JAG12279.1 GEBQ01013060 GEBQ01010336 JAT26917.1 JAT29641.1 GBHO01012041 GBHO01012038 JAG31563.1 JAG31566.1 LBMM01010997 KMQ87090.1 DMQF01000352 HAO15199.1 GEBQ01027915 JAT12062.1 GBBI01004799 JAC13913.1 DMQF01000361 HAO15230.1 DNUT01000507 HBI41157.1 GEHC01000896 JAV46749.1 GANO01004126 JAB55745.1 LBMM01004152 KMQ92759.1 DMQF01000157 HAO14486.1 DNUT01000056 HBI39512.1 DMQF01000003 HAO13943.1 DNUT01000451 HBI40968.1 GANO01003893 JAB55978.1 GANO01004094 JAB55777.1 GAMC01021038 GAMC01021033 GAMC01021031 JAB85522.1 GAMC01021037 GAMC01021036 JAB85519.1 QOIP01000004 RLU23301.1 GANO01004066 JAB55805.1 GANO01004361 JAB55510.1 DMQF01000478 HAO15711.1 GANO01004338 JAB55533.1 GEBQ01030598 JAT09379.1 GANO01004362 JAB55509.1 GANO01004198 JAB55673.1 DNUT01000170 HBI39961.1 GEZM01074146 JAV64657.1 GANO01004238 JAB55633.1 LSMT01000554 PFX16333.1 GEHC01001007 JAV46638.1 GEHC01000892 JAV46753.1 GANO01004275 JAB55596.1 DMQF01000030 HAO14052.1 GEHC01000940 JAV46705.1 GANO01003995 JAB55876.1 GEHC01000909 JAV46736.1 GEZM01041318 GEZM01041317 JAV80524.1 GALA01001237 JAA93615.1 DNUT01000297 HBI40414.1 GEZM01074147 JAV64656.1 GEZM01007604 JAV95063.1 AAZX01004038 GECU01021424 JAS86282.1 GANO01004070 JAB55801.1 GANO01004325 JAB55546.1 GANO01004166 JAB55705.1 GANO01001936 JAB57935.1 AAZX01023386 GEZM01008907 JAV94684.1 GEZM01000581 JAV98361.1 NWSH01001817 PCG70032.1 GEZM01051381 GEZM01051380 JAV74995.1 GFDF01007660 JAV06424.1 KZ150070 PZC74075.1 GEZM01045590 GEZM01045589 JAV77734.1 NWSH01000010 PCG80923.1 GANO01004069 JAB55802.1 GBHO01015041 JAG28563.1 AAZX01004360 AAZX01016008 AAZX01017248 DQ099730 AAZ15237.1 GANO01004258 JAB55613.1 LBMM01003452 KMQ93522.1 LSMT01000038 PFX31271.1 NWSH01000998 PCG73165.1 LSMT01000382 PFX18982.1
LBMM01014048 KMQ85356.1 LBMM01017805 KMQ83966.1 GBHO01031326 GBHO01031325 JAG12278.1 JAG12279.1 GEBQ01013060 GEBQ01010336 JAT26917.1 JAT29641.1 GBHO01012041 GBHO01012038 JAG31563.1 JAG31566.1 LBMM01010997 KMQ87090.1 DMQF01000352 HAO15199.1 GEBQ01027915 JAT12062.1 GBBI01004799 JAC13913.1 DMQF01000361 HAO15230.1 DNUT01000507 HBI41157.1 GEHC01000896 JAV46749.1 GANO01004126 JAB55745.1 LBMM01004152 KMQ92759.1 DMQF01000157 HAO14486.1 DNUT01000056 HBI39512.1 DMQF01000003 HAO13943.1 DNUT01000451 HBI40968.1 GANO01003893 JAB55978.1 GANO01004094 JAB55777.1 GAMC01021038 GAMC01021033 GAMC01021031 JAB85522.1 GAMC01021037 GAMC01021036 JAB85519.1 QOIP01000004 RLU23301.1 GANO01004066 JAB55805.1 GANO01004361 JAB55510.1 DMQF01000478 HAO15711.1 GANO01004338 JAB55533.1 GEBQ01030598 JAT09379.1 GANO01004362 JAB55509.1 GANO01004198 JAB55673.1 DNUT01000170 HBI39961.1 GEZM01074146 JAV64657.1 GANO01004238 JAB55633.1 LSMT01000554 PFX16333.1 GEHC01001007 JAV46638.1 GEHC01000892 JAV46753.1 GANO01004275 JAB55596.1 DMQF01000030 HAO14052.1 GEHC01000940 JAV46705.1 GANO01003995 JAB55876.1 GEHC01000909 JAV46736.1 GEZM01041318 GEZM01041317 JAV80524.1 GALA01001237 JAA93615.1 DNUT01000297 HBI40414.1 GEZM01074147 JAV64656.1 GEZM01007604 JAV95063.1 AAZX01004038 GECU01021424 JAS86282.1 GANO01004070 JAB55801.1 GANO01004325 JAB55546.1 GANO01004166 JAB55705.1 GANO01001936 JAB57935.1 AAZX01023386 GEZM01008907 JAV94684.1 GEZM01000581 JAV98361.1 NWSH01001817 PCG70032.1 GEZM01051381 GEZM01051380 JAV74995.1 GFDF01007660 JAV06424.1 KZ150070 PZC74075.1 GEZM01045590 GEZM01045589 JAV77734.1 NWSH01000010 PCG80923.1 GANO01004069 JAB55802.1 GBHO01015041 JAG28563.1 AAZX01004360 AAZX01016008 AAZX01017248 DQ099730 AAZ15237.1 GANO01004258 JAB55613.1 LBMM01003452 KMQ93522.1 LSMT01000038 PFX31271.1 NWSH01000998 PCG73165.1 LSMT01000382 PFX18982.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JRM8
A0A1B6L6Y3
A0A2A4JPY6
A0A0J7K589
A0A0J7K195
A0A0A9WX05
+ More
A0A1B6LTE5 A0A0A9YIX2 A0A0J7N335 A0A3C1S1N3 A0A1B6KKS7 A0A023EXE1 A0A3C1S2P1 A0A354GJG9 A0A1W7R6F6 U5EQQ2 A0A0J7KR96 A0A3C1RZS6 A0A354GES4 A0A3C1RY27 A0A354GIY0 A0A1S3DPM6 U5EUG4 U5ENG8 W8AIM4 W8B6W6 A0A3L8DSB3 U5EQV9 U5EQB9 A0A3C1S360 U5EQA6 A0A1B6KD49 U5EEC7 U5EN99 A0A354GG23 A0A1Y1L0L4 U5ETR7 A0A2B4RHP5 A0A1W7R648 A0A1W7R6J3 U5EEL4 A0A3C1RYE8 A0A1W7R6K8 U5ER05 A0A1W7R6E4 A0A1Y1M3U5 T1DE06 A0A354GHC6 A0A1Y1KZ78 A0A1Y1NB24 K7JTK2 A0A1B6IH82 U5EQT8 U5EQE2 U5ETW8 U5EVC6 K7JAP1 A0A1Y1N9S9 A0A1Y1NKD6 A0A2A4JF59 A0A1Y1LSZ2 A0A1L8DJB0 A0A2W1BK42 A0A1Y1LVZ3 A0A3B3HNZ2 A0A2A4KAV4 U5EF12 A0A0A9YBT8 A0A3B3BNA3 K7JBA8 Q4F8Q2 U5EQJ2 A0A0J7KSZ7 A0A2B4SN01 A0A2A4JM54 A0A3B3HIF3 A0A2B4RRX2
A0A1B6LTE5 A0A0A9YIX2 A0A0J7N335 A0A3C1S1N3 A0A1B6KKS7 A0A023EXE1 A0A3C1S2P1 A0A354GJG9 A0A1W7R6F6 U5EQQ2 A0A0J7KR96 A0A3C1RZS6 A0A354GES4 A0A3C1RY27 A0A354GIY0 A0A1S3DPM6 U5EUG4 U5ENG8 W8AIM4 W8B6W6 A0A3L8DSB3 U5EQV9 U5EQB9 A0A3C1S360 U5EQA6 A0A1B6KD49 U5EEC7 U5EN99 A0A354GG23 A0A1Y1L0L4 U5ETR7 A0A2B4RHP5 A0A1W7R648 A0A1W7R6J3 U5EEL4 A0A3C1RYE8 A0A1W7R6K8 U5ER05 A0A1W7R6E4 A0A1Y1M3U5 T1DE06 A0A354GHC6 A0A1Y1KZ78 A0A1Y1NB24 K7JTK2 A0A1B6IH82 U5EQT8 U5EQE2 U5ETW8 U5EVC6 K7JAP1 A0A1Y1N9S9 A0A1Y1NKD6 A0A2A4JF59 A0A1Y1LSZ2 A0A1L8DJB0 A0A2W1BK42 A0A1Y1LVZ3 A0A3B3HNZ2 A0A2A4KAV4 U5EF12 A0A0A9YBT8 A0A3B3BNA3 K7JBA8 Q4F8Q2 U5EQJ2 A0A0J7KSZ7 A0A2B4SN01 A0A2A4JM54 A0A3B3HIF3 A0A2B4RRX2
PDB
6AR3
E-value=1.42254e-06,
Score=127
Ontologies
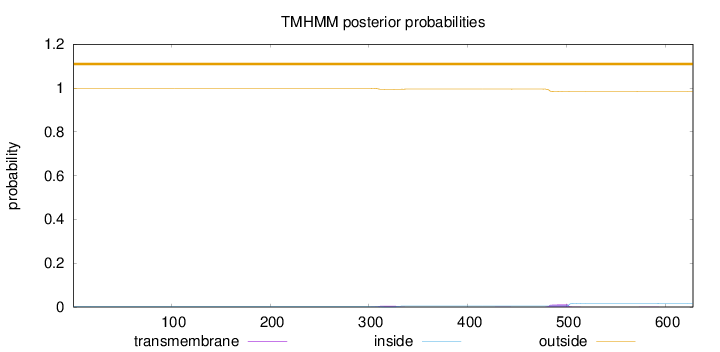
Topology
Length:
628
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.38629
Exp number, first 60 AAs:
0.00117
Total prob of N-in:
0.00149
outside
1 - 628
Population Genetic Test Statistics
Pi
16.908168
Theta
14.406706
Tajima's D
0.520761
CLR
0
CSRT
0.526423678816059
Interpretation
Uncertain
