Gene
KWMTBOMO03192
Pre Gene Modal
BGIBMGA013646
Annotation
PREDICTED:_iodotyrosine_dehalogenase_1_[Papilio_xuthus]
Full name
Iodotyrosine deiodinase 1
Alternative Name
Iodotyrosine dehalogenase 1
Location in the cell
Nuclear Reliability : 1.943
Sequence
CDS
ATGAATATTATATCGCTAGTTGTTTGGACATTGGAGAAGAACTGTTTTATCGTGTACACAGCATTAATAGTTTGTTTTATAGGCTTCACGATATTCCACAAGAACAGATTGGATATTCATTACAGGAATGTTGAAATAAAGGAAGAAAAATTGGCGCGAACCGGCCCAAAGAAAAAAGAACGTGAATTCCCTGATGATGGGGACGATGACCCAGAGTTACTCCTGCCAGCAATCCCGGAAGACACTCCGCACATACCTTACCGACCTCCGAGGAAATCCGATGAAGAAATCCTTCAGAAGTCCAGGGAATATTATGAGCTGATGGCCAAAAGGCGAACGGTCAGATGTTTTAGCGCTGAGCCTATACCTCAGGAAGTCTTGGATAATATTGTGAAGACTGCTGGCACTGCACCATCAGGCGCTCACACTGAACCCTGGACCTTTGTAGTGGTCCAAGATCCACAAATTAAGGAAGCAATCCGCGAGATTGTAGAACAAGAAGAGGAATTAAACTACACCCAGAGGATGTCCAGACAATGGGTGACTGATCTCAAACCTTTTGCCACAAAGCCAAGCAAACCTTACCTATCTGAAGCACCAGCTTTAGTTCTGGTGTTCAGACAGGCGTACTCCTGGAGATCAGATGGAAAGAAACGGATGCATTACTATAGTGAGATCAGCACGGCCATCGCAGCAGGATTTCTGCTAGCAGCTATTCAGTACTGCGGCCTAGTGGCGCTTACATCGACACCTCTAAATGCCAACGCTCGTCTCCGGGATCTCCTCTCGAGACCAGCCAATGAGAGACTGGAACTCCTTCTGCCGGTAGGAAGGCCGCACAAGGAAACAACTGTACCTGATTTAGATAGAAAGCCTTTAGAAGAGATCATGCTTAAGATCTGA
Protein
MNIISLVVWTLEKNCFIVYTALIVCFIGFTIFHKNRLDIHYRNVEIKEEKLARTGPKKKEREFPDDGDDDPELLLPAIPEDTPHIPYRPPRKSDEEILQKSREYYELMAKRRTVRCFSAEPIPQEVLDNIVKTAGTAPSGAHTEPWTFVVVQDPQIKEAIREIVEQEEELNYTQRMSRQWVTDLKPFATKPSKPYLSEAPALVLVFRQAYSWRSDGKKRMHYYSEISTAIAAGFLLAAIQYCGLVALTSTPLNANARLRDLLSRPANERLELLLPVGRPHKETTVPDLDRKPLEEIMLKI
Summary
Description
Catalyzes the oxidative NADPH-dependent deiodination of monoiodotyrosine (L-MIT) or diiodotyrosine (L-DIT). Acts during the hydrolysis of thyroglobulin to liberate iodide, which can then reenter the hormone-producing pathways. Acts more efficiently on monoiodotyrosine than on diiodotyrosine.
Catalyzes the oxidative NADPH-dependent deiodination of monoiodotyrosine (L-MIT) or diiodotyrosine (L-DIT) (PubMed:15289438, PubMed:25395621, PubMed:18434651). Acts during the hydrolysis of thyroglobulin to liberate iodide, which can then reenter the hormone-producing pathways (PubMed:18434651). Acts more efficiently on monoiodotyrosine than on diiodotyrosine (PubMed:15289438).
Catalyzes the oxidative NADPH-dependent deiodination of monoiodotyrosine (L-MIT) or diiodotyrosine (L-DIT) (PubMed:15289438, PubMed:25395621, PubMed:18434651). Acts during the hydrolysis of thyroglobulin to liberate iodide, which can then reenter the hormone-producing pathways (PubMed:18434651). Acts more efficiently on monoiodotyrosine than on diiodotyrosine (PubMed:15289438).
Catalytic Activity
2 iodide + L-tyrosine + 2 NADP(+) = 3,5-diiodo-L-tyrosine + H(+) + 2 NADPH
Cofactor
FMN
Biophysicochemical Properties
2.67 uM for L-DIT
1.35 uM for L-MIT
1.35 uM for L-MIT
Subunit
Homodimer.
Similarity
Belongs to the nitroreductase family.
Keywords
Cell membrane
Complete proteome
Cytoplasmic vesicle
Flavoprotein
FMN
Membrane
NADP
Oxidoreductase
Reference proteome
Signal
Transmembrane
Transmembrane helix
3D-structure
Alternative splicing
Congenital hypothyroidism
Disease mutation
Polymorphism
Feature
chain Iodotyrosine deiodinase 1
splice variant In isoform 7.
sequence variant In TDH4; strongly reduces activity; does not respond to the increase of flavin mononucleotide concentration; dbSNP:rs121918138.
splice variant In isoform 7.
sequence variant In TDH4; strongly reduces activity; does not respond to the increase of flavin mononucleotide concentration; dbSNP:rs121918138.
Uniprot
A0A212F7F3
A0A194Q2F6
A0A0N0PCT3
A0A2A4JJG2
E0VY32
A0A023F693
+ More
A0A0A9VZ71 U6CRC6 A0A067QI15 N6TUM2 A0A1U7SH75 U4UEI0 F7G6J3 M3YG65 T2MBC4 S9WYP1 A0A1S3DTX9 A0A2Y9KZB1 Q5M8L2 A0A2J7RFY6 G1PBC7 A0A224XUI5 G3VRG0 S7NPQ6 E2R3F8 A0A1D2N124 W5NH61 A0A384BBU3 L5KFR3 A0A2U4CNL6 A0A2K6GN80 A0A2K6THK6 Q5REW1 A0A151NM03 A0A2K5DS00 H2PKM1 A0A2Y9MA64 A0A340XBK5 A0A341BND5 W4YXJ6 A0A093IFE1 A0A2K5SD20 A0A2I3GFV1 A0A2K5NU14 Q6PHW0 A0A096MYS1 A0A384CFM7 A0A091I4X7 A0A3B4UGH1 A0A2K6RRY2 A0A093R224 A0A195ES38 A0A2K6JTE7 A0A091P7U2 A0A0Q3US12 A0A2I2YAN4 A0A287BQ51 H0WUU8 A0A1D5Q4F0 A0A2R8M9R6 A0A2K6CKT5 H9GBT3 E2AVW2 A0A093K8Y2 A0A2Y9HHK8 A0A2K5YZJ4 A0A0F8AVE2 A0A2Y9ELN2 E1C0X6 A0A2K5UT53 A0A2D4HYH2 A0A091T2Q0 Q66KT6 E9IKN4 A0A3B4WUT9 A0A2K5IXZ5 A0A1L8G7Q2 A0A091W310 A0A093DQL0 A0A2R9B550 M3W0V6 A0A091G6B1 A0A2U3W7E9 A0A3B5AS70 A0A094L5D4 A0A2I3TW58 A0A2I0MTL1 F4W5M4 A0A091KIQ3 A0A091NDG1 A0A2D0SI30 A0A1A8EIP4 A0A091T8E8 A0A3P9K2U3 A0A226PUQ9
A0A0A9VZ71 U6CRC6 A0A067QI15 N6TUM2 A0A1U7SH75 U4UEI0 F7G6J3 M3YG65 T2MBC4 S9WYP1 A0A1S3DTX9 A0A2Y9KZB1 Q5M8L2 A0A2J7RFY6 G1PBC7 A0A224XUI5 G3VRG0 S7NPQ6 E2R3F8 A0A1D2N124 W5NH61 A0A384BBU3 L5KFR3 A0A2U4CNL6 A0A2K6GN80 A0A2K6THK6 Q5REW1 A0A151NM03 A0A2K5DS00 H2PKM1 A0A2Y9MA64 A0A340XBK5 A0A341BND5 W4YXJ6 A0A093IFE1 A0A2K5SD20 A0A2I3GFV1 A0A2K5NU14 Q6PHW0 A0A096MYS1 A0A384CFM7 A0A091I4X7 A0A3B4UGH1 A0A2K6RRY2 A0A093R224 A0A195ES38 A0A2K6JTE7 A0A091P7U2 A0A0Q3US12 A0A2I2YAN4 A0A287BQ51 H0WUU8 A0A1D5Q4F0 A0A2R8M9R6 A0A2K6CKT5 H9GBT3 E2AVW2 A0A093K8Y2 A0A2Y9HHK8 A0A2K5YZJ4 A0A0F8AVE2 A0A2Y9ELN2 E1C0X6 A0A2K5UT53 A0A2D4HYH2 A0A091T2Q0 Q66KT6 E9IKN4 A0A3B4WUT9 A0A2K5IXZ5 A0A1L8G7Q2 A0A091W310 A0A093DQL0 A0A2R9B550 M3W0V6 A0A091G6B1 A0A2U3W7E9 A0A3B5AS70 A0A094L5D4 A0A2I3TW58 A0A2I0MTL1 F4W5M4 A0A091KIQ3 A0A091NDG1 A0A2D0SI30 A0A1A8EIP4 A0A091T8E8 A0A3P9K2U3 A0A226PUQ9
EC Number
1.21.1.1
Pubmed
22118469
26354079
20566863
25474469
25401762
24845553
+ More
23537049 17495919 24065732 23149746 20431018 21993624 21709235 16341006 27289101 23258410 22293439 14574404 15489334 14702039 15289438 24275569 25395621 18434651 24813606 25362486 22398555 30723633 17431167 21881562 20798317 25835551 15592404 21282665 27762356 22722832 17975172 16136131 23371554 21719571 17554307 24621616
23537049 17495919 24065732 23149746 20431018 21993624 21709235 16341006 27289101 23258410 22293439 14574404 15489334 14702039 15289438 24275569 25395621 18434651 24813606 25362486 22398555 30723633 17431167 21881562 20798317 25835551 15592404 21282665 27762356 22722832 17975172 16136131 23371554 21719571 17554307 24621616
EMBL
AGBW02009879
OWR49660.1
KQ459579
KPI99518.1
KQ460434
KPJ14760.1
+ More
NWSH01001314 PCG71734.1 DS235843 EEB18288.1 GBBI01001965 JAC16747.1 GBHO01044126 JAF99477.1 HAAF01001225 CCP73051.1 KK853366 KDR08112.1 APGK01008157 APGK01018039 KB740064 KB737617 ENN81768.1 ENN82987.1 KB632132 ERL89011.1 AEYP01034373 HAAD01003154 CDG69386.1 KB017067 EPY80644.1 AAMC01122984 BC087975 AAH87975.1 NEVH01004408 PNF39738.1 AAPE02047741 GFTR01004815 JAW11611.1 AEFK01173337 AEFK01173338 AEFK01173339 AEFK01173340 KE164616 EPQ19221.1 AAEX03000269 LJIJ01000310 ODM98938.1 AHAT01015800 KB030829 ELK09338.1 CR857405 AKHW03002600 KYO37699.1 ABGA01293022 ABGA01293023 ABGA01293024 ABGA01293025 NDHI03003369 PNJ78989.1 AAGJ04097708 KK586712 KFV97493.1 ADFV01166569 ADFV01166570 ADFV01166571 AY259176 AY259177 AY424901 AY424902 AY957659 AY957660 AY957661 AL031010 BC056253 AK129950 AHZZ02025568 KL218240 KFP03272.1 KL425797 KFW90227.1 KQ981993 KYN31043.1 KK653785 KFQ03461.1 LMAW01002567 KQK79802.1 CABD030048437 AEMK02000001 DQIR01168408 DQIR01293609 HDB23885.1 AAQR03140332 JSUE03031928 AAWZ02022304 GL443213 EFN62384.1 KL206904 KFV86844.1 KQ040945 KKF32702.1 AADN05000003 AQIA01050252 IACK01070805 LAA77003.1 KK495778 KFQ65492.1 BC078565 AAH78565.1 GL764021 EFZ18868.1 CM004474 OCT79893.1 KK734675 KFR10017.1 KL449365 KFV04446.1 AJFE02036324 AJFE02036325 AANG04000169 KL447083 KFO69732.1 KL272168 KFZ64647.1 AACZ04040565 NBAG03000056 PNI93106.1 AKCR02000002 PKK33016.1 GL887655 EGI70521.1 KK741999 KFP39055.1 KK845149 KFP86924.1 HAEB01000528 SBQ46976.1 KK442700 KFQ70719.1 AWGT02000010 OXB83761.1
NWSH01001314 PCG71734.1 DS235843 EEB18288.1 GBBI01001965 JAC16747.1 GBHO01044126 JAF99477.1 HAAF01001225 CCP73051.1 KK853366 KDR08112.1 APGK01008157 APGK01018039 KB740064 KB737617 ENN81768.1 ENN82987.1 KB632132 ERL89011.1 AEYP01034373 HAAD01003154 CDG69386.1 KB017067 EPY80644.1 AAMC01122984 BC087975 AAH87975.1 NEVH01004408 PNF39738.1 AAPE02047741 GFTR01004815 JAW11611.1 AEFK01173337 AEFK01173338 AEFK01173339 AEFK01173340 KE164616 EPQ19221.1 AAEX03000269 LJIJ01000310 ODM98938.1 AHAT01015800 KB030829 ELK09338.1 CR857405 AKHW03002600 KYO37699.1 ABGA01293022 ABGA01293023 ABGA01293024 ABGA01293025 NDHI03003369 PNJ78989.1 AAGJ04097708 KK586712 KFV97493.1 ADFV01166569 ADFV01166570 ADFV01166571 AY259176 AY259177 AY424901 AY424902 AY957659 AY957660 AY957661 AL031010 BC056253 AK129950 AHZZ02025568 KL218240 KFP03272.1 KL425797 KFW90227.1 KQ981993 KYN31043.1 KK653785 KFQ03461.1 LMAW01002567 KQK79802.1 CABD030048437 AEMK02000001 DQIR01168408 DQIR01293609 HDB23885.1 AAQR03140332 JSUE03031928 AAWZ02022304 GL443213 EFN62384.1 KL206904 KFV86844.1 KQ040945 KKF32702.1 AADN05000003 AQIA01050252 IACK01070805 LAA77003.1 KK495778 KFQ65492.1 BC078565 AAH78565.1 GL764021 EFZ18868.1 CM004474 OCT79893.1 KK734675 KFR10017.1 KL449365 KFV04446.1 AJFE02036324 AJFE02036325 AANG04000169 KL447083 KFO69732.1 KL272168 KFZ64647.1 AACZ04040565 NBAG03000056 PNI93106.1 AKCR02000002 PKK33016.1 GL887655 EGI70521.1 KK741999 KFP39055.1 KK845149 KFP86924.1 HAEB01000528 SBQ46976.1 KK442700 KFQ70719.1 AWGT02000010 OXB83761.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000218220
UP000009046
UP000027135
+ More
UP000019118 UP000189705 UP000030742 UP000002280 UP000000715 UP000079169 UP000248482 UP000008143 UP000235965 UP000001074 UP000007648 UP000002254 UP000094527 UP000018468 UP000261681 UP000010552 UP000245320 UP000233160 UP000233220 UP000001595 UP000050525 UP000233020 UP000248483 UP000265300 UP000252040 UP000007110 UP000233040 UP000001073 UP000233060 UP000005640 UP000028761 UP000261680 UP000291021 UP000054308 UP000261420 UP000233200 UP000078541 UP000233180 UP000051836 UP000001519 UP000008227 UP000005225 UP000006718 UP000008225 UP000233120 UP000001646 UP000000311 UP000053584 UP000248481 UP000233140 UP000248484 UP000000539 UP000233100 UP000261360 UP000233080 UP000186698 UP000053605 UP000240080 UP000011712 UP000053760 UP000245340 UP000261400 UP000002277 UP000053872 UP000007755 UP000221080 UP000265180 UP000198419
UP000019118 UP000189705 UP000030742 UP000002280 UP000000715 UP000079169 UP000248482 UP000008143 UP000235965 UP000001074 UP000007648 UP000002254 UP000094527 UP000018468 UP000261681 UP000010552 UP000245320 UP000233160 UP000233220 UP000001595 UP000050525 UP000233020 UP000248483 UP000265300 UP000252040 UP000007110 UP000233040 UP000001073 UP000233060 UP000005640 UP000028761 UP000261680 UP000291021 UP000054308 UP000261420 UP000233200 UP000078541 UP000233180 UP000051836 UP000001519 UP000008227 UP000005225 UP000006718 UP000008225 UP000233120 UP000001646 UP000000311 UP000053584 UP000248481 UP000233140 UP000248484 UP000000539 UP000233100 UP000261360 UP000233080 UP000186698 UP000053605 UP000240080 UP000011712 UP000053760 UP000245340 UP000261400 UP000002277 UP000053872 UP000007755 UP000221080 UP000265180 UP000198419
Pfam
PF00881 Nitroreductase
SUPFAM
SSF55469
SSF55469
Gene 3D
ProteinModelPortal
A0A212F7F3
A0A194Q2F6
A0A0N0PCT3
A0A2A4JJG2
E0VY32
A0A023F693
+ More
A0A0A9VZ71 U6CRC6 A0A067QI15 N6TUM2 A0A1U7SH75 U4UEI0 F7G6J3 M3YG65 T2MBC4 S9WYP1 A0A1S3DTX9 A0A2Y9KZB1 Q5M8L2 A0A2J7RFY6 G1PBC7 A0A224XUI5 G3VRG0 S7NPQ6 E2R3F8 A0A1D2N124 W5NH61 A0A384BBU3 L5KFR3 A0A2U4CNL6 A0A2K6GN80 A0A2K6THK6 Q5REW1 A0A151NM03 A0A2K5DS00 H2PKM1 A0A2Y9MA64 A0A340XBK5 A0A341BND5 W4YXJ6 A0A093IFE1 A0A2K5SD20 A0A2I3GFV1 A0A2K5NU14 Q6PHW0 A0A096MYS1 A0A384CFM7 A0A091I4X7 A0A3B4UGH1 A0A2K6RRY2 A0A093R224 A0A195ES38 A0A2K6JTE7 A0A091P7U2 A0A0Q3US12 A0A2I2YAN4 A0A287BQ51 H0WUU8 A0A1D5Q4F0 A0A2R8M9R6 A0A2K6CKT5 H9GBT3 E2AVW2 A0A093K8Y2 A0A2Y9HHK8 A0A2K5YZJ4 A0A0F8AVE2 A0A2Y9ELN2 E1C0X6 A0A2K5UT53 A0A2D4HYH2 A0A091T2Q0 Q66KT6 E9IKN4 A0A3B4WUT9 A0A2K5IXZ5 A0A1L8G7Q2 A0A091W310 A0A093DQL0 A0A2R9B550 M3W0V6 A0A091G6B1 A0A2U3W7E9 A0A3B5AS70 A0A094L5D4 A0A2I3TW58 A0A2I0MTL1 F4W5M4 A0A091KIQ3 A0A091NDG1 A0A2D0SI30 A0A1A8EIP4 A0A091T8E8 A0A3P9K2U3 A0A226PUQ9
A0A0A9VZ71 U6CRC6 A0A067QI15 N6TUM2 A0A1U7SH75 U4UEI0 F7G6J3 M3YG65 T2MBC4 S9WYP1 A0A1S3DTX9 A0A2Y9KZB1 Q5M8L2 A0A2J7RFY6 G1PBC7 A0A224XUI5 G3VRG0 S7NPQ6 E2R3F8 A0A1D2N124 W5NH61 A0A384BBU3 L5KFR3 A0A2U4CNL6 A0A2K6GN80 A0A2K6THK6 Q5REW1 A0A151NM03 A0A2K5DS00 H2PKM1 A0A2Y9MA64 A0A340XBK5 A0A341BND5 W4YXJ6 A0A093IFE1 A0A2K5SD20 A0A2I3GFV1 A0A2K5NU14 Q6PHW0 A0A096MYS1 A0A384CFM7 A0A091I4X7 A0A3B4UGH1 A0A2K6RRY2 A0A093R224 A0A195ES38 A0A2K6JTE7 A0A091P7U2 A0A0Q3US12 A0A2I2YAN4 A0A287BQ51 H0WUU8 A0A1D5Q4F0 A0A2R8M9R6 A0A2K6CKT5 H9GBT3 E2AVW2 A0A093K8Y2 A0A2Y9HHK8 A0A2K5YZJ4 A0A0F8AVE2 A0A2Y9ELN2 E1C0X6 A0A2K5UT53 A0A2D4HYH2 A0A091T2Q0 Q66KT6 E9IKN4 A0A3B4WUT9 A0A2K5IXZ5 A0A1L8G7Q2 A0A091W310 A0A093DQL0 A0A2R9B550 M3W0V6 A0A091G6B1 A0A2U3W7E9 A0A3B5AS70 A0A094L5D4 A0A2I3TW58 A0A2I0MTL1 F4W5M4 A0A091KIQ3 A0A091NDG1 A0A2D0SI30 A0A1A8EIP4 A0A091T8E8 A0A3P9K2U3 A0A226PUQ9
PDB
4TTC
E-value=1.0992e-64,
Score=624
Ontologies
GO
Topology
Subcellular location
Cell membrane
Cytoplasmic vesicle membrane
Cytoplasmic vesicle membrane
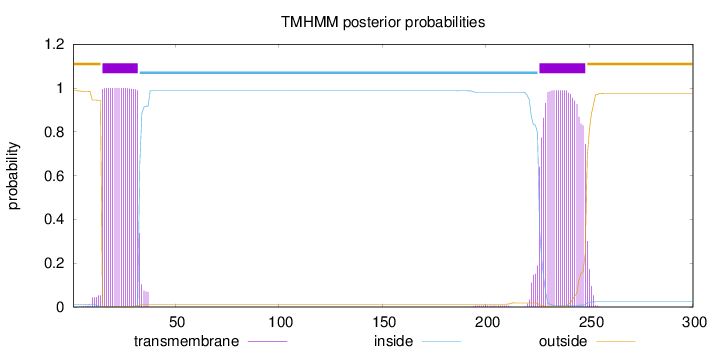
Length:
300
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.42152
Exp number, first 60 AAs:
18.88122
Total prob of N-in:
0.01259
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 32
inside
33 - 225
TMhelix
226 - 248
outside
249 - 300
Population Genetic Test Statistics
Pi
177.661443
Theta
141.512551
Tajima's D
-1.189524
CLR
0.544196
CSRT
0.104844757762112
Interpretation
Uncertain
