Gene
KWMTBOMO03182
Pre Gene Modal
BGIBMGA013453
Annotation
cyclin-dependent_kinase_2_[Bombyx_mori]
Full name
Cyclin-dependent kinase 2
Alternative Name
Cell division protein kinase 2
p33 protein kinase
p33 protein kinase
Location in the cell
Cytoplasmic Reliability : 2.383
Sequence
CDS
ATGGAGAATTTTTCTACTGTAGAAAAAATAGGGGAAGGAACATATGGTGTAGTTTACAAAGCCAAAGACAGGGTCACCGGACAAGAAATCGCGTTGAAGAAAATCAAACTTGAAAATGAACCTGAGGGTGTACCATCAACAGCCTTACGGGAGATCTCAGTGCTGCGAGAACTTCGACATCCAGCAGTTGTACGGCTCCTTGATGTTATGCTGGCATCTTCAGACAGTAAACTGTTTCTTGTCTTTGAATATTTGAATATGGATCTCAAAAGACTCATGGATCTGACGAAGGGACCACTACCTATTGATTTAGTTAAGAGTTATTTACGACAGTTACTGGAAGGTGTTGCGTACTGTCACGCCCAACGTGTTCTCCACAGAGACCTGAAGCCCCAGAACCTACTAATAGACGAGGAAGGTCACATTAAATTGGCCGATTTCGGTTTGGCTCGAGCTTTTGGTATCCCTGTGCGTGCTTACACACATGAGGTGGTCACACTATGGTACCGTGCACCAGAAATCCTTCTTGGGGCTAAGTTCTACTCAACCGCCGTTGATGTTTGGAGCTTAGCCTGTATTTTTGCAGAGATGGCAAGCGGTCGCACTCTTTTCCCCGGTGACAGTGAAATAGACCAGCTATTCCGCGTATTCCGAGCGCTGGGTACACCAGGCGAGGCACTATGGCCGGCAGCGAGAAGACTTCCTGACTTCCGAGCCGCCTTCCCGCGATGGCCTGCCCGACCTGCCCGAACATTACTACCAGCAGGACTCCGAGCGCACAGTTCAGCGGCAGCGTTGTTTGAGGCCATGCTGCGTTATGAACCTGAGACTCGAATTCCTGCCCGAGCTGCTCTTACGCATCCTTACTTGGCTGATGCGACTCTGGTGCCGCCTCCTCTACATCCTTAA
Protein
MENFSTVEKIGEGTYGVVYKAKDRVTGQEIALKKIKLENEPEGVPSTALREISVLRELRHPAVVRLLDVMLASSDSKLFLVFEYLNMDLKRLMDLTKGPLPIDLVKSYLRQLLEGVAYCHAQRVLHRDLKPQNLLIDEEGHIKLADFGLARAFGIPVRAYTHEVVTLWYRAPEILLGAKFYSTAVDVWSLACIFAEMASGRTLFPGDSEIDQLFRVFRALGTPGEALWPAARRLPDFRAAFPRWPARPARTLLPAGLRAHSSAAALFEAMLRYEPETRIPARAALTHPYLADATLVPPPLHP
Summary
Description
Serine/threonine-protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis. Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, EZH2. Triggers duplication of centrosomes and DNA. Acts at the G1-S transition to promote the E2F transcriptional program and the initiation of DNA synthesis, and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation, and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in human embryonic stem cells (hESCs). Activity of CDK2 is maximal during S phase and G2; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1-S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1 (By similarity). Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1-S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress towards mitosis. In response to DNA damage, double-strand break repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociates from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1-S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the nitric oxide- (NO) mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1-S transition. CTNNB1 phosphorylation regulates insulin internalization. Phosphorylates FOXP3 and negatively regulates its transcriptional activity and protein stability (PubMed:23853094). Phosphorylates CDK2AP2 (By similarity).
Serine/threonine-protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis. Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, EZH2. Triggers duplication of centrosomes and DNA. Acts at the G1-S transition to promote the E2F transcriptional program and the initiation of DNA synthesis, and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation, and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in human embryonic stem cells (hESCs). Activity of CDK2 is maximal during S phase and G2; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1-S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1 (By similarity). Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1-S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress towards mitosis. In response to DNA damage, double-strand break repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociates from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1-S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the nitric oxide- (NO) mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1-S transition. CTNNB1 phosphorylation regulates insulin internalization. Phosphorylates FOXP3 and negatively regulates its transcriptional activity and protein stability (By similarity). Phosphorylates CDK2AP2 (By similarity).
Serine/threonine-protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis. Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, EZH2. Triggers duplication of centrosomes and DNA. Acts at the G1-S transition to promote the E2F transcriptional program and the initiation of DNA synthesis, and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation, and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in human embryonic stem cells (hESCs). Activity of CDK2 is maximal during S phase and G2; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1-S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1 (By similarity). Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1-S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress towards mitosis. In response to DNA damage, double-strand break repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociates from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1-S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the nitric oxide- (NO) mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1-S transition. CTNNB1 phosphorylation regulates insulin internalization. Phosphorylates FOXP3 and negatively regulates its transcriptional activity and protein stability (By similarity). Phosphorylates CDK2AP2 (PubMed:12944431).
Serine/threonine-protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis. Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, EZH2. Triggers duplication of centrosomes and DNA. Acts at the G1-S transition to promote the E2F transcriptional program and the initiation of DNA synthesis, and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation, and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in human embryonic stem cells (hESCs). Activity of CDK2 is maximal during S phase and G2; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1-S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1 (By similarity). Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1-S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress towards mitosis. In response to DNA damage, double-strand break repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociates from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1-S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the nitric oxide- (NO) mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1-S transition. CTNNB1 phosphorylation regulates insulin internalization. Phosphorylates FOXP3 and negatively regulates its transcriptional activity and protein stability (By similarity). Phosphorylates CDK2AP2 (By similarity).
Serine/threonine-protein kinase involved in the control of the cell cycle; essential for meiosis, but dispensable for mitosis. Phosphorylates CTNNB1, USP37, p53/TP53, NPM1, CDK7, RB1, BRCA2, MYC, NPAT, EZH2. Triggers duplication of centrosomes and DNA. Acts at the G1-S transition to promote the E2F transcriptional program and the initiation of DNA synthesis, and modulates G2 progression; controls the timing of entry into mitosis/meiosis by controlling the subsequent activation of cyclin B/CDK1 by phosphorylation, and coordinates the activation of cyclin B/CDK1 at the centrosome and in the nucleus. Crucial role in orchestrating a fine balance between cellular proliferation, cell death, and DNA repair in human embryonic stem cells (hESCs). Activity of CDK2 is maximal during S phase and G2; activated by interaction with cyclin E during the early stages of DNA synthesis to permit G1-S transition, and subsequently activated by cyclin A2 (cyclin A1 in germ cells) during the late stages of DNA replication to drive the transition from S phase to mitosis, the G2 phase. EZH2 phosphorylation promotes H3K27me3 maintenance and epigenetic gene silencing. Phosphorylates CABLES1 (By similarity). Cyclin E/CDK2 prevents oxidative stress-mediated Ras-induced senescence by phosphorylating MYC. Involved in G1-S phase DNA damage checkpoint that prevents cells with damaged DNA from initiating mitosis; regulates homologous recombination-dependent repair by phosphorylating BRCA2, this phosphorylation is low in S phase when recombination is active, but increases as cells progress towards mitosis. In response to DNA damage, double-strand break repair by homologous recombination a reduction of CDK2-mediated BRCA2 phosphorylation. Phosphorylation of RB1 disturbs its interaction with E2F1. NPM1 phosphorylation by cyclin E/CDK2 promotes its dissociates from unduplicated centrosomes, thus initiating centrosome duplication. Cyclin E/CDK2-mediated phosphorylation of NPAT at G1-S transition and until prophase stimulates the NPAT-mediated activation of histone gene transcription during S phase. Required for vitamin D-mediated growth inhibition by being itself inactivated. Involved in the nitric oxide- (NO) mediated signaling in a nitrosylation/activation-dependent manner. USP37 is activated by phosphorylation and thus triggers G1-S transition. CTNNB1 phosphorylation regulates insulin internalization. Phosphorylates FOXP3 and negatively regulates its transcriptional activity and protein stability (By similarity). Phosphorylates CDK2AP2 (PubMed:12944431).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Subunit
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1 (PubMed:11585773). Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC. Interaction with SPDYA promotes kinase activation via a conformation change that alleviates obstruction of the substrate-binding cleft by the T-loop. Found in a complex with both SPDYA and CDKN1B/KIP1. Binds to RB1 and CDK7. Binding to CDKN1A (p21) leads to CDK2/cyclin E inactivation at the G1-S phase DNA damage checkpoint, thereby arresting cells at the G1-S transition during DNA repair. Associated with PTPN6 and beta-catenin/CTNNB1. Interacts with CACUL1. May interact with CEP63. Interacts with ANKRD17 (By similarity). Interacts with CEBPA (when phosphorylated) (PubMed:15107404). Forms a ternary complex with CCNA2 and CDKN1B; CDKN1B inhibits the kinase activity of CDK2 through conformational rearrangements. Interacts with cyclins A, B1, B3, D, or E. Interacts with CDK2AP2 (By similarity).
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1 (By similarity). Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC. Interaction with SPDYA promotes kinase activation via a conformation change that alleviates obstruction of the substrate-binding cleft by the T-loop. Found in a complex with both SPDYA and CDKN1B/KIP1. Binds to RB1 (PubMed:10542199). Binds to CDK7. Binding to CDKN1A (p21) leads to CDK2/cyclin E inactivation at the G1-S phase DNA damage checkpoint, thereby arresting cells at the G1-S transition during DNA repair. Associated with PTPN6 and beta-catenin/CTNNB1. Interacts with CACUL1. May interact with CEP63. Interacts with ANKRD17. Interacts with CEBPA (when phosphorylated). Forms a ternary complex with CCNA2 and CDKN1B; CDKN1B inhibits the kinase activity of CDK2 through conformational rearrangements. Interacts with cyclins A, B1, B3, D, or E. Interacts with CDK2AP2 (By similarity).
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1 (By similarity). Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC. Interaction with SPDYA promotes kinase activation via a conformation change that alleviates obstruction of the substrate-binding cleft by the T-loop. Found in a complex with both SPDYA and CDKN1B/KIP1. Binds to RB1 and CDK7. Binding to CDKN1A (p21) leads to CDK2/cyclin E inactivation at the G1-S phase DNA damage checkpoint, thereby arresting cells at the G1-S transition during DNA repair. Associated with PTPN6 and beta-catenin/CTNNB1. Interacts with CACUL1. May interact with CEP63. Interacts with ANKRD17. Interacts with CEBPA (when phosphorylated). Forms a ternary complex with CCNA2 and CDKN1B; CDKN1B inhibits the kinase activity of CDK2 through conformational rearrangements. Interacts with cyclins A, B1, B3, D, or E. Interacts with CDK2AP2 (By similarity).
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1 (By similarity). Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC (PubMed:15611625). Interaction with SPDYA promotes kinase activation via a conformation change that alleviates obstruction of the substrate-binding cleft by the T-loop (PubMed:28666995). Found in a complex with both SPDYA and CDKN1B/KIP1 (PubMed:12972555, PubMed:28666995). Binds to RB1 and CDK7. Binding to CDKN1A (p21) leads to CDK2/cyclin E inactivation at the G1-S phase DNA damage checkpoint, thereby arresting cells at the G1-S transition during DNA repair. Associated with PTPN6 and beta-catenin/CTNNB1. Interacts with CACUL1. May interact with CEP63. Interacts with ANKRD17. Interacts with CEBPA (when phosphorylated) (PubMed:15107404). Forms a ternary complex with CCNA2 and CDKN1B; CDKN1B inhibits the kinase activity of CDK2 through conformational rearrangements (PubMed:8684460). Interacts with cyclins A, B1, B3, D, or E (PubMed:10499802, PubMed:10884347, PubMed:12185076, PubMed:23781148). Interacts with CDK2AP2 (PubMed:23781148).
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1 (By similarity). Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC. Interaction with SPDYA promotes kinase activation via a conformation change that alleviates obstruction of the substrate-binding cleft by the T-loop. Found in a complex with both SPDYA and CDKN1B/KIP1. Binds to RB1 (PubMed:10542199). Binds to CDK7. Binding to CDKN1A (p21) leads to CDK2/cyclin E inactivation at the G1-S phase DNA damage checkpoint, thereby arresting cells at the G1-S transition during DNA repair. Associated with PTPN6 and beta-catenin/CTNNB1. Interacts with CACUL1. May interact with CEP63. Interacts with ANKRD17. Interacts with CEBPA (when phosphorylated). Forms a ternary complex with CCNA2 and CDKN1B; CDKN1B inhibits the kinase activity of CDK2 through conformational rearrangements. Interacts with cyclins A, B1, B3, D, or E. Interacts with CDK2AP2 (By similarity).
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1 (By similarity). Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC. Interaction with SPDYA promotes kinase activation via a conformation change that alleviates obstruction of the substrate-binding cleft by the T-loop. Found in a complex with both SPDYA and CDKN1B/KIP1. Binds to RB1 and CDK7. Binding to CDKN1A (p21) leads to CDK2/cyclin E inactivation at the G1-S phase DNA damage checkpoint, thereby arresting cells at the G1-S transition during DNA repair. Associated with PTPN6 and beta-catenin/CTNNB1. Interacts with CACUL1. May interact with CEP63. Interacts with ANKRD17. Interacts with CEBPA (when phosphorylated). Forms a ternary complex with CCNA2 and CDKN1B; CDKN1B inhibits the kinase activity of CDK2 through conformational rearrangements. Interacts with cyclins A, B1, B3, D, or E. Interacts with CDK2AP2 (By similarity).
Found in a complex with CABLES1, CCNA1 and CCNE1. Interacts with CABLES1 (By similarity). Interacts with UHRF2. Part of a complex consisting of UHRF2, CDK2 and CCNE1. Interacts with the Speedy/Ringo proteins SPDYA and SPDYC (PubMed:15611625). Interaction with SPDYA promotes kinase activation via a conformation change that alleviates obstruction of the substrate-binding cleft by the T-loop (PubMed:28666995). Found in a complex with both SPDYA and CDKN1B/KIP1 (PubMed:12972555, PubMed:28666995). Binds to RB1 and CDK7. Binding to CDKN1A (p21) leads to CDK2/cyclin E inactivation at the G1-S phase DNA damage checkpoint, thereby arresting cells at the G1-S transition during DNA repair. Associated with PTPN6 and beta-catenin/CTNNB1. Interacts with CACUL1. May interact with CEP63. Interacts with ANKRD17. Interacts with CEBPA (when phosphorylated) (PubMed:15107404). Forms a ternary complex with CCNA2 and CDKN1B; CDKN1B inhibits the kinase activity of CDK2 through conformational rearrangements (PubMed:8684460). Interacts with cyclins A, B1, B3, D, or E (PubMed:10499802, PubMed:10884347, PubMed:12185076, PubMed:23781148). Interacts with CDK2AP2 (PubMed:23781148).
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Keywords
Acetylation
Alternative splicing
ATP-binding
Cell cycle
Cell division
Complete proteome
Cytoplasm
Cytoskeleton
DNA damage
DNA repair
Endosome
Kinase
Magnesium
Meiosis
Metal-binding
Mitosis
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Transferase
3D-structure
Polymorphism
S-nitrosylation
Feature
chain Cyclin-dependent kinase 2
splice variant In isoform CDK2-alpha.
sequence variant In dbSNP:rs3087335.
splice variant In isoform CDK2-alpha.
sequence variant In dbSNP:rs3087335.
Uniprot
H6VMF4
A0A2H1V8G8
S4PMI6
A0A437BI96
A0A2W1BA74
A0A2A4JVV4
+ More
A0A212ELY3 A0A0L7LM44 A0A0N0U5C0 H3BI57 O46161 A0A2A3E5Z8 A0A087ZNC2 A0A310SE63 G1P317 S7NB09 V4B3L0 A0A0L7R4I8 A0A154PB06 G3TH75 W4XW48 T0MEU0 A0A287AXP1 A0A2Y9GUJ7 F6US96 E2QW70 A0A341AR07 A0A3Q7RF90 A0A340Y2B2 A0A2I2V4K7 A0A2U3XBV5 A0A2Y9L521 A0A3Q7SQ67 A0A2Y9NEE7 A0A2U3W1B4 A0A383ZXG1 A0A452RHR8 A0A3Q7X0F6 A0A384DTG0 D2H336 H0WVH2 A0A2Y9FRU8 A0A2K6GY84 A0A1S3FMH0 P97377-2 Q6P751 Q3U6X7 A0A2Y9DS22 L8Y724 Q63699 G3W161 E2ALN7 I3LWC0 T1J0E3 A0A2K5DF40 A0A2K5QXK8 F7BWH8 A0A1U8C8E6 O55076 A0A286XH18 G5BB95 L8HYF4 B7U171 A0MSV8 Q5E9Y0 A0A401PCX1 A0A1U7U8H5 A0A2K6E4W9 A0A2K6MMD4 G3QIR0 A0A2K5XCR8 A0A0D9QZ04 A0A2R9B4E4 A0A096MYH8 A0A2K6P9I2 A0A2K5NT27 A0A2K5IHJ1 H2NHM8 G7PIG0 G7N7A7 H2Q670 A0A024RB77 P24941 A0A158NMZ5 A0A2Y9GC92 A2IAR9 A0A151JVM2 A0A3M6URQ2 P48963 M3W4N5 A0A452GT32 A0A2Y9GF67 A0A226MDZ0 A0A226MMP6 A0A195DBS4 F1RW06 A0A151I170 A0A401SB05 H9GA76 A0A1S3ALH3
A0A212ELY3 A0A0L7LM44 A0A0N0U5C0 H3BI57 O46161 A0A2A3E5Z8 A0A087ZNC2 A0A310SE63 G1P317 S7NB09 V4B3L0 A0A0L7R4I8 A0A154PB06 G3TH75 W4XW48 T0MEU0 A0A287AXP1 A0A2Y9GUJ7 F6US96 E2QW70 A0A341AR07 A0A3Q7RF90 A0A340Y2B2 A0A2I2V4K7 A0A2U3XBV5 A0A2Y9L521 A0A3Q7SQ67 A0A2Y9NEE7 A0A2U3W1B4 A0A383ZXG1 A0A452RHR8 A0A3Q7X0F6 A0A384DTG0 D2H336 H0WVH2 A0A2Y9FRU8 A0A2K6GY84 A0A1S3FMH0 P97377-2 Q6P751 Q3U6X7 A0A2Y9DS22 L8Y724 Q63699 G3W161 E2ALN7 I3LWC0 T1J0E3 A0A2K5DF40 A0A2K5QXK8 F7BWH8 A0A1U8C8E6 O55076 A0A286XH18 G5BB95 L8HYF4 B7U171 A0MSV8 Q5E9Y0 A0A401PCX1 A0A1U7U8H5 A0A2K6E4W9 A0A2K6MMD4 G3QIR0 A0A2K5XCR8 A0A0D9QZ04 A0A2R9B4E4 A0A096MYH8 A0A2K6P9I2 A0A2K5NT27 A0A2K5IHJ1 H2NHM8 G7PIG0 G7N7A7 H2Q670 A0A024RB77 P24941 A0A158NMZ5 A0A2Y9GC92 A2IAR9 A0A151JVM2 A0A3M6URQ2 P48963 M3W4N5 A0A452GT32 A0A2Y9GF67 A0A226MDZ0 A0A226MMP6 A0A195DBS4 F1RW06 A0A151I170 A0A401SB05 H9GA76 A0A1S3ALH3
EC Number
2.7.11.22
Pubmed
19121390
23622113
28756777
22118469
26227816
9215903
+ More
9705226 21993624 23254933 23149746 23836398 19892987 16341006 17975172 24813606 20010809 15489334 11585773 11733001 14561402 12923533 15107404 17942597 21183079 23853094 15057822 15632090 10349636 11042159 11076861 11217851 12466851 12040188 16141073 23385571 7862443 8673024 10542199 21709235 20798317 25243066 11506705 21993625 22751099 19340603 16305752 30297745 22398555 22722832 25362486 22002653 25319552 16136131 11181995 1714386 1653904 1717994 14702039 16541075 1396589 9030781 10499802 11051553 10995386 10995387 10884347 11113184 12185076 11980914 12839962 12944431 12972555 15178429 14597612 15611625 15800615 17373709 18691976 18372919 19829063 19369195 19690332 19608861 20147522 20079829 20360007 20935635 20195506 19966300 21269460 21406398 21262353 21684737 21596315 21319273 19445729 19238148 19561645 20713526 21517772 22814378 23781148 8510751 7630397 8601310 8917641 8684460 8756328 8610110 9334743 9677190 16325401 17570665 17495531 17095507 17937404 18656911 21565702 28666995 17344846 23711367 21347285 15592404 17635936 30382153 8280171 28562605 24621616 21881562
9705226 21993624 23254933 23149746 23836398 19892987 16341006 17975172 24813606 20010809 15489334 11585773 11733001 14561402 12923533 15107404 17942597 21183079 23853094 15057822 15632090 10349636 11042159 11076861 11217851 12466851 12040188 16141073 23385571 7862443 8673024 10542199 21709235 20798317 25243066 11506705 21993625 22751099 19340603 16305752 30297745 22398555 22722832 25362486 22002653 25319552 16136131 11181995 1714386 1653904 1717994 14702039 16541075 1396589 9030781 10499802 11051553 10995386 10995387 10884347 11113184 12185076 11980914 12839962 12944431 12972555 15178429 14597612 15611625 15800615 17373709 18691976 18372919 19829063 19369195 19690332 19608861 20147522 20079829 20360007 20935635 20195506 19966300 21269460 21406398 21262353 21684737 21596315 21319273 19445729 19238148 19561645 20713526 21517772 22814378 23781148 8510751 7630397 8601310 8917641 8684460 8756328 8610110 9334743 9677190 16325401 17570665 17495531 17095507 17937404 18656911 21565702 28666995 17344846 23711367 21347285 15592404 17635936 30382153 8280171 28562605 24621616 21881562
EMBL
BABH01037322
JN833622
AFB82433.1
ODYU01001044
SOQ36702.1
GAIX01003535
+ More
JAA89025.1 RSAL01000053 RVE50053.1 KZ150187 PZC72439.1 NWSH01000543 PCG75764.1 AGBW02013968 OWR42504.1 JTDY01000669 KOB76276.1 KQ435789 KOX74501.1 AFYH01001758 AFYH01001759 AJ224917 CAA12223.1 KZ288357 PBC27130.1 KQ760761 OAD59142.1 AAPE02026874 KE163877 EPQ14231.1 KB203827 ESO82954.1 KQ414657 KOC65758.1 KQ434850 KZC08564.1 AAGJ04171849 KB016734 EQB77761.1 AEMK02000033 JX967576 AGR86101.1 AAEX03006916 AANG04003655 ACTA01178059 GL192457 EFB28834.1 AAQR03076828 AAQR03076829 U63337 AJ223732 AJ223733 BC005654 AC098012 AC141508 BC061832 CH474104 AAH61832.1 EDL84822.1 AK152922 CH466578 BAE31597.1 EDL24600.1 KB364768 ELV12032.1 D28753 D63162 AEFK01203592 GL440609 EFN65646.1 AGTP01085299 AGTP01085300 JH431734 GAMT01001273 GAMS01008093 GAMQ01007798 JAB10588.1 JAB15043.1 JAB34053.1 AJ223949 AAKN02014842 JH169329 GEBF01002266 EHB06556.1 JAO01367.1 JH882551 ELR48983.1 FJ422550 ACJ53943.1 LWLT01000005 EF035041 ABK34941.1 BT020790 BC150026 BFAA01003391 GCB70959.1 CABD030083632 AQIB01023022 AJFE02087845 AHZZ02002258 ABGA01042610 ABGA01042611 NDHI03003457 PNJ44889.1 AQIA01011136 CM001286 EHH66383.1 JU320958 JU473899 JV045857 CM001263 AFE64714.1 AFH30703.1 AFI35928.1 EHH20842.1 AACZ04012961 GABC01007850 GABF01000440 GABD01001183 GABE01004554 NBAG03000092 JAA03488.1 JAA21705.1 JAA31917.1 JAA40185.1 PNI90081.1 CH471054 EAW96860.1 X61622 X62071 M68520 AB012305 BT006821 AF512553 AK291941 AC025162 AC034102 BC003065 ADTU01020711 ADTU01020712 AADN05000742 EF182713 ABM55710.1 KQ981689 KYN37898.1 RCHS01000860 RMX56356.1 D17350 AANG04004184 MCFN01001318 OXB53259.1 AWGT02008165 OXB56521.1 KQ981010 KYN10338.1 AEMK02000080 KQ976593 KYM79677.1 BEZZ01000169 GCC27553.1 AAWZ02011909
JAA89025.1 RSAL01000053 RVE50053.1 KZ150187 PZC72439.1 NWSH01000543 PCG75764.1 AGBW02013968 OWR42504.1 JTDY01000669 KOB76276.1 KQ435789 KOX74501.1 AFYH01001758 AFYH01001759 AJ224917 CAA12223.1 KZ288357 PBC27130.1 KQ760761 OAD59142.1 AAPE02026874 KE163877 EPQ14231.1 KB203827 ESO82954.1 KQ414657 KOC65758.1 KQ434850 KZC08564.1 AAGJ04171849 KB016734 EQB77761.1 AEMK02000033 JX967576 AGR86101.1 AAEX03006916 AANG04003655 ACTA01178059 GL192457 EFB28834.1 AAQR03076828 AAQR03076829 U63337 AJ223732 AJ223733 BC005654 AC098012 AC141508 BC061832 CH474104 AAH61832.1 EDL84822.1 AK152922 CH466578 BAE31597.1 EDL24600.1 KB364768 ELV12032.1 D28753 D63162 AEFK01203592 GL440609 EFN65646.1 AGTP01085299 AGTP01085300 JH431734 GAMT01001273 GAMS01008093 GAMQ01007798 JAB10588.1 JAB15043.1 JAB34053.1 AJ223949 AAKN02014842 JH169329 GEBF01002266 EHB06556.1 JAO01367.1 JH882551 ELR48983.1 FJ422550 ACJ53943.1 LWLT01000005 EF035041 ABK34941.1 BT020790 BC150026 BFAA01003391 GCB70959.1 CABD030083632 AQIB01023022 AJFE02087845 AHZZ02002258 ABGA01042610 ABGA01042611 NDHI03003457 PNJ44889.1 AQIA01011136 CM001286 EHH66383.1 JU320958 JU473899 JV045857 CM001263 AFE64714.1 AFH30703.1 AFI35928.1 EHH20842.1 AACZ04012961 GABC01007850 GABF01000440 GABD01001183 GABE01004554 NBAG03000092 JAA03488.1 JAA21705.1 JAA31917.1 JAA40185.1 PNI90081.1 CH471054 EAW96860.1 X61622 X62071 M68520 AB012305 BT006821 AF512553 AK291941 AC025162 AC034102 BC003065 ADTU01020711 ADTU01020712 AADN05000742 EF182713 ABM55710.1 KQ981689 KYN37898.1 RCHS01000860 RMX56356.1 D17350 AANG04004184 MCFN01001318 OXB53259.1 AWGT02008165 OXB56521.1 KQ981010 KYN10338.1 AEMK02000080 KQ976593 KYM79677.1 BEZZ01000169 GCC27553.1 AAWZ02011909
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000037510
UP000053105
+ More
UP000008672 UP000242457 UP000005203 UP000001074 UP000030746 UP000053825 UP000076502 UP000007646 UP000007110 UP000008227 UP000248481 UP000002281 UP000002254 UP000252040 UP000286641 UP000265300 UP000011712 UP000245341 UP000248482 UP000286640 UP000248483 UP000245340 UP000261681 UP000291022 UP000286642 UP000261680 UP000291021 UP000008912 UP000005225 UP000248484 UP000233160 UP000081671 UP000000589 UP000002494 UP000248480 UP000011518 UP000007648 UP000000311 UP000005215 UP000233020 UP000233040 UP000008225 UP000189706 UP000005447 UP000006813 UP000291000 UP000009136 UP000288216 UP000189704 UP000233120 UP000233180 UP000001519 UP000233140 UP000029965 UP000240080 UP000028761 UP000233200 UP000233060 UP000233080 UP000001595 UP000009130 UP000233100 UP000002277 UP000005640 UP000005205 UP000000539 UP000078541 UP000275408 UP000291020 UP000198323 UP000198419 UP000078492 UP000078540 UP000287033 UP000001646 UP000079721
UP000008672 UP000242457 UP000005203 UP000001074 UP000030746 UP000053825 UP000076502 UP000007646 UP000007110 UP000008227 UP000248481 UP000002281 UP000002254 UP000252040 UP000286641 UP000265300 UP000011712 UP000245341 UP000248482 UP000286640 UP000248483 UP000245340 UP000261681 UP000291022 UP000286642 UP000261680 UP000291021 UP000008912 UP000005225 UP000248484 UP000233160 UP000081671 UP000000589 UP000002494 UP000248480 UP000011518 UP000007648 UP000000311 UP000005215 UP000233020 UP000233040 UP000008225 UP000189706 UP000005447 UP000006813 UP000291000 UP000009136 UP000288216 UP000189704 UP000233120 UP000233180 UP000001519 UP000233140 UP000029965 UP000240080 UP000028761 UP000233200 UP000233060 UP000233080 UP000001595 UP000009130 UP000233100 UP000002277 UP000005640 UP000005205 UP000000539 UP000078541 UP000275408 UP000291020 UP000198323 UP000198419 UP000078492 UP000078540 UP000287033 UP000001646 UP000079721
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H6VMF4
A0A2H1V8G8
S4PMI6
A0A437BI96
A0A2W1BA74
A0A2A4JVV4
+ More
A0A212ELY3 A0A0L7LM44 A0A0N0U5C0 H3BI57 O46161 A0A2A3E5Z8 A0A087ZNC2 A0A310SE63 G1P317 S7NB09 V4B3L0 A0A0L7R4I8 A0A154PB06 G3TH75 W4XW48 T0MEU0 A0A287AXP1 A0A2Y9GUJ7 F6US96 E2QW70 A0A341AR07 A0A3Q7RF90 A0A340Y2B2 A0A2I2V4K7 A0A2U3XBV5 A0A2Y9L521 A0A3Q7SQ67 A0A2Y9NEE7 A0A2U3W1B4 A0A383ZXG1 A0A452RHR8 A0A3Q7X0F6 A0A384DTG0 D2H336 H0WVH2 A0A2Y9FRU8 A0A2K6GY84 A0A1S3FMH0 P97377-2 Q6P751 Q3U6X7 A0A2Y9DS22 L8Y724 Q63699 G3W161 E2ALN7 I3LWC0 T1J0E3 A0A2K5DF40 A0A2K5QXK8 F7BWH8 A0A1U8C8E6 O55076 A0A286XH18 G5BB95 L8HYF4 B7U171 A0MSV8 Q5E9Y0 A0A401PCX1 A0A1U7U8H5 A0A2K6E4W9 A0A2K6MMD4 G3QIR0 A0A2K5XCR8 A0A0D9QZ04 A0A2R9B4E4 A0A096MYH8 A0A2K6P9I2 A0A2K5NT27 A0A2K5IHJ1 H2NHM8 G7PIG0 G7N7A7 H2Q670 A0A024RB77 P24941 A0A158NMZ5 A0A2Y9GC92 A2IAR9 A0A151JVM2 A0A3M6URQ2 P48963 M3W4N5 A0A452GT32 A0A2Y9GF67 A0A226MDZ0 A0A226MMP6 A0A195DBS4 F1RW06 A0A151I170 A0A401SB05 H9GA76 A0A1S3ALH3
A0A212ELY3 A0A0L7LM44 A0A0N0U5C0 H3BI57 O46161 A0A2A3E5Z8 A0A087ZNC2 A0A310SE63 G1P317 S7NB09 V4B3L0 A0A0L7R4I8 A0A154PB06 G3TH75 W4XW48 T0MEU0 A0A287AXP1 A0A2Y9GUJ7 F6US96 E2QW70 A0A341AR07 A0A3Q7RF90 A0A340Y2B2 A0A2I2V4K7 A0A2U3XBV5 A0A2Y9L521 A0A3Q7SQ67 A0A2Y9NEE7 A0A2U3W1B4 A0A383ZXG1 A0A452RHR8 A0A3Q7X0F6 A0A384DTG0 D2H336 H0WVH2 A0A2Y9FRU8 A0A2K6GY84 A0A1S3FMH0 P97377-2 Q6P751 Q3U6X7 A0A2Y9DS22 L8Y724 Q63699 G3W161 E2ALN7 I3LWC0 T1J0E3 A0A2K5DF40 A0A2K5QXK8 F7BWH8 A0A1U8C8E6 O55076 A0A286XH18 G5BB95 L8HYF4 B7U171 A0MSV8 Q5E9Y0 A0A401PCX1 A0A1U7U8H5 A0A2K6E4W9 A0A2K6MMD4 G3QIR0 A0A2K5XCR8 A0A0D9QZ04 A0A2R9B4E4 A0A096MYH8 A0A2K6P9I2 A0A2K5NT27 A0A2K5IHJ1 H2NHM8 G7PIG0 G7N7A7 H2Q670 A0A024RB77 P24941 A0A158NMZ5 A0A2Y9GC92 A2IAR9 A0A151JVM2 A0A3M6URQ2 P48963 M3W4N5 A0A452GT32 A0A2Y9GF67 A0A226MDZ0 A0A226MMP6 A0A195DBS4 F1RW06 A0A151I170 A0A401SB05 H9GA76 A0A1S3ALH3
Ontologies
KEGG
GO
GO:0004674
GO:0005524
GO:0051301
GO:0019904
GO:0008284
GO:0097134
GO:0000781
GO:0005768
GO:0030332
GO:0060968
GO:0000287
GO:0005667
GO:0000805
GO:0032298
GO:0000122
GO:0007265
GO:0035173
GO:0000806
GO:0007099
GO:0005813
GO:0015030
GO:0045893
GO:0018105
GO:0097124
GO:0006813
GO:0004693
GO:0000082
GO:0000793
GO:0097123
GO:0097135
GO:0005634
GO:0000307
GO:0006468
GO:0005737
GO:0010389
GO:0016572
GO:0006281
GO:0044877
GO:0097472
GO:0043231
GO:0005654
GO:0005829
GO:0032869
GO:0004672
GO:0051321
GO:0051591
GO:0016301
GO:0051602
GO:0046686
GO:0045471
GO:0005815
GO:0046872
GO:0032355
GO:0002088
GO:0031100
GO:0042493
GO:0009636
GO:0030496
GO:0000086
GO:0071732
GO:0006260
GO:0006977
GO:0031571
GO:0031145
GO:0051298
GO:1901796
GO:0006974
GO:0003697
GO:1990879
GO:0046982
GO:0005615
GO:0005515
GO:0016021
GO:0045211
GO:0048384
Topology
Subcellular location
Cytoplasm
Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Cytoskeleton Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Microtubule organizing center Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Centrosome Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Nucleus Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Cajal body Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Endosome Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Cytoskeleton Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Microtubule organizing center Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Centrosome Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Nucleus Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Cajal body Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
Endosome Localized at the centrosomes in late G2 phase after separation of the centrosomes but before the start of prophase. Nuclear-cytoplasmic trafficking is mediated during the inhibition by 1,25-(OH)(2)D(3) (By similarity). With evidence from 2 publications.
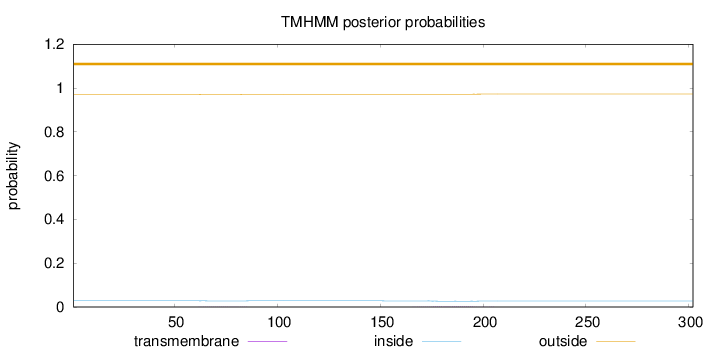
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0643299999999999
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.02863
outside
1 - 302
Population Genetic Test Statistics
Pi
40.983936
Theta
33.996669
Tajima's D
-2.236398
CLR
1.164567
CSRT
0.00454977251137443
Interpretation
Uncertain
