Gene
KWMTBOMO03180
Annotation
PREDICTED:_unconventional_myosin-Ia-like_isoform_X2_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 2.296
Sequence
CDS
ATGGCGGAGGGCGAAGCGACAGGTGTAGCAGACGCGGTGCTCCTGGCACCACTTACAGAGGACACGTTTCTGCACAACCTGCATGTCCGCTTCAAGCGTGATGTTATTTATACCTATGTTGGCAATGCGTTAATTTCGGTGAATCCATGCCGGCCCTTGCCACTCTACTCGGCAGACCTGGTGCGTGCCTATCTCGCCCGACCTCCATACACGTTGCCCCCACATTTATATGCAATAACAGTGACCGCTTACCGTTGGGTGCGAGATAGAAACGAAAGTCAATGCATAGTGATTACAGGTTGGAACATTACTTTTGTCTTTCTGTTAGCATAA
Protein
MAEGEATGVADAVLLAPLTEDTFLHNLHVRFKRDVIYTYVGNALISVNPCRPLPLYSADLVRAYLARPPYTLPPHLYAITVTAYRWVRDRNESQCIVITGWNITFVFLLA
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
A0A2W1BDI3
A0A2A4JWJ4
A0A2A4JUS6
A0A2A4JWI6
A0A2H1V8X2
A0A194Q2K8
+ More
A0A212ELY4 A0A0N1PI54 A0A1Y1K1E7 A0A3Q0IYS9 D6WBX6 U4UKC3 N6U416 A0A1B6HVB5 E0VD23 T1HXQ9 V4CKW9 A0A2P8XZ55 A0A1B6DN56 A0A0S7JXX8 A0A3Q2P298 A0A402G3X2 A0A210PRF8 A0A0B7BA75 A0A0B7B9V2 A0A0B7B8Z7 A0A0B7BCD9 B7Q0V0 A0A3B3T082 A0A3B3T034 C9JUP5 A0A2J8N4H2 A0A2J8WVQ8 C9JYW1 F6W622 A0A1W2WAC1 F6W6J2 A0A2C9MAE6 A0A2J8N4H0 A0A2J8WVS1 W4XUC0 A0A3P8P7F9 A0A2T7PV62 S4R706 F7G232 E7EQD9 K1RK57 Q05CR5 A0A2P4S4B9 H2ZWQ3 A0A1W4ZDJ7 R7TWD0 A0A1W4Z4U8 A0A1W4ZE34 A0A1W4ZDH8 A0A1W4Z4U7 A0A1W4ZDJ2 D3YV80 A0A1S3HRA3 A0A1S3HQK3 H2Z8V8 H2Z8V7 A7RGF6 A0A1S3PNB1 A0A0S7JW73 A0A2D4HNJ1 V9KQF7 A0A3B4CMM8 A0A3B4E642 A0A3B4E7L7 A0A3B4E6D9 A0A0S7JWJ0 A0A401T467 A0A3B3UL06 A0A3B3XM75 A0A3B5PU45 A0A087XEB4 A0A3B3UKB7 A0A3P9PM21 A0A3P9PM46 A0A3B5QR98 A0A3P9PM01 A0A3P9PLM7 A0A2I4BW27 A0A2D4HNF6 M4AQ39 A0A3P9PLF5 A0A2Y9L955 A0A2Y9L8Y0 A0A2Y9LED4 A0A3S3NY11 A0A2I0MJ07 W5MA88 A0A146VHI7 A0A146VIH2 A0A146VHH4 A0A1V4K708 A0A1V4K6Y0 A0A3Q3GGA1 A0A3Q3AZD4
A0A212ELY4 A0A0N1PI54 A0A1Y1K1E7 A0A3Q0IYS9 D6WBX6 U4UKC3 N6U416 A0A1B6HVB5 E0VD23 T1HXQ9 V4CKW9 A0A2P8XZ55 A0A1B6DN56 A0A0S7JXX8 A0A3Q2P298 A0A402G3X2 A0A210PRF8 A0A0B7BA75 A0A0B7B9V2 A0A0B7B8Z7 A0A0B7BCD9 B7Q0V0 A0A3B3T082 A0A3B3T034 C9JUP5 A0A2J8N4H2 A0A2J8WVQ8 C9JYW1 F6W622 A0A1W2WAC1 F6W6J2 A0A2C9MAE6 A0A2J8N4H0 A0A2J8WVS1 W4XUC0 A0A3P8P7F9 A0A2T7PV62 S4R706 F7G232 E7EQD9 K1RK57 Q05CR5 A0A2P4S4B9 H2ZWQ3 A0A1W4ZDJ7 R7TWD0 A0A1W4Z4U8 A0A1W4ZE34 A0A1W4ZDH8 A0A1W4Z4U7 A0A1W4ZDJ2 D3YV80 A0A1S3HRA3 A0A1S3HQK3 H2Z8V8 H2Z8V7 A7RGF6 A0A1S3PNB1 A0A0S7JW73 A0A2D4HNJ1 V9KQF7 A0A3B4CMM8 A0A3B4E642 A0A3B4E7L7 A0A3B4E6D9 A0A0S7JWJ0 A0A401T467 A0A3B3UL06 A0A3B3XM75 A0A3B5PU45 A0A087XEB4 A0A3B3UKB7 A0A3P9PM21 A0A3P9PM46 A0A3B5QR98 A0A3P9PM01 A0A3P9PLM7 A0A2I4BW27 A0A2D4HNF6 M4AQ39 A0A3P9PLF5 A0A2Y9L955 A0A2Y9L8Y0 A0A2Y9LED4 A0A3S3NY11 A0A2I0MJ07 W5MA88 A0A146VHI7 A0A146VIH2 A0A146VHH4 A0A1V4K708 A0A1V4K6Y0 A0A3Q3GGA1 A0A3Q3AZD4
Pubmed
28756777
26354079
22118469
28004739
18362917
19820115
+ More
23537049 20566863 23254933 29403074 28812685 29240929 15815621 24275569 12481130 15114417 15562597 18464734 21269460 25218447 25114211 25755297 28112733 22992520 15489334 9215903 19468303 21183079 17615350 24402279 30297745 23542700 23371554
23537049 20566863 23254933 29403074 28812685 29240929 15815621 24275569 12481130 15114417 15562597 18464734 21269460 25218447 25114211 25755297 28112733 22992520 15489334 9215903 19468303 21183079 17615350 24402279 30297745 23542700 23371554
EMBL
KZ150187
PZC72441.1
NWSH01000543
PCG75762.1
PCG75761.1
PCG75760.1
+ More
ODYU01001044 SOQ36704.1 KQ459579 KPI99568.1 AGBW02013968 OWR42502.1 KQ459753 KPJ20180.1 GEZM01099740 GEZM01099739 GEZM01099737 GEZM01099734 JAV53166.1 KQ971316 EEZ98757.2 KB632237 ERL90445.1 APGK01043658 KB741015 ENN75366.1 GECU01029092 JAS78614.1 DS235070 EEB11279.1 ACPB03004606 KB200129 ESP02895.1 PYGN01001136 PSN37289.1 GEDC01010226 JAS27072.1 GBYX01227683 JAO69459.1 BDOT01028337 GCF63389.1 NEDP02005545 OWF39079.1 HACG01042943 HACG01042947 CEK89808.1 CEK89812.1 HACG01042948 HACG01042949 CEK89813.1 CEK89814.1 HACG01042942 HACG01042944 CEK89807.1 CEK89809.1 HACG01042945 HACG01042946 CEK89810.1 CEK89811.1 ABJB010345952 ABJB010358880 ABJB010602649 ABJB010640774 ABJB010643890 ABJB010720724 ABJB010733083 ABJB010780283 ABJB010960659 ABJB011052605 DS834001 EEC12472.1 AC068056 AC092614 AC118061 AC133534 NBAG03000236 PNI66665.1 NDHI03003377 PNJ73859.1 PNI66669.1 PNJ73858.1 AAGJ04102190 AAGJ04102191 AAGJ04102192 PZQS01000002 PVD37309.1 JH818444 EKC42000.1 BC021665 AAH21665.1 PPHD01112613 POI18964.1 AFYH01211787 AFYH01211788 AFYH01211789 AFYH01211790 AFYH01211791 AFYH01211792 AFYH01211793 AFYH01211794 AFYH01211795 AFYH01211796 AMQN01010558 KB308144 ELT98223.1 AC108840 AC123555 AC138680 DS469509 EDO49515.1 GBYX01227677 JAO69464.1 IACK01050379 LAA73508.1 JW867793 AFP00311.1 GBYX01227690 GBYX01227688 GBYX01227684 GBYX01227680 JAO69454.1 BEZZ01000995 GCC37407.1 AYCK01009418 IACK01050380 LAA73509.1 NCKU01003092 RWS08190.1 AKCR02000010 PKK29661.1 AHAT01011349 AHAT01011350 AHAT01011351 AHAT01011352 AHAT01011353 AHAT01011354 AHAT01011355 GCES01069483 JAR16840.1 GCES01069484 JAR16839.1 GCES01069482 JAR16841.1 LSYS01004331 OPJ80175.1 OPJ80174.1
ODYU01001044 SOQ36704.1 KQ459579 KPI99568.1 AGBW02013968 OWR42502.1 KQ459753 KPJ20180.1 GEZM01099740 GEZM01099739 GEZM01099737 GEZM01099734 JAV53166.1 KQ971316 EEZ98757.2 KB632237 ERL90445.1 APGK01043658 KB741015 ENN75366.1 GECU01029092 JAS78614.1 DS235070 EEB11279.1 ACPB03004606 KB200129 ESP02895.1 PYGN01001136 PSN37289.1 GEDC01010226 JAS27072.1 GBYX01227683 JAO69459.1 BDOT01028337 GCF63389.1 NEDP02005545 OWF39079.1 HACG01042943 HACG01042947 CEK89808.1 CEK89812.1 HACG01042948 HACG01042949 CEK89813.1 CEK89814.1 HACG01042942 HACG01042944 CEK89807.1 CEK89809.1 HACG01042945 HACG01042946 CEK89810.1 CEK89811.1 ABJB010345952 ABJB010358880 ABJB010602649 ABJB010640774 ABJB010643890 ABJB010720724 ABJB010733083 ABJB010780283 ABJB010960659 ABJB011052605 DS834001 EEC12472.1 AC068056 AC092614 AC118061 AC133534 NBAG03000236 PNI66665.1 NDHI03003377 PNJ73859.1 PNI66669.1 PNJ73858.1 AAGJ04102190 AAGJ04102191 AAGJ04102192 PZQS01000002 PVD37309.1 JH818444 EKC42000.1 BC021665 AAH21665.1 PPHD01112613 POI18964.1 AFYH01211787 AFYH01211788 AFYH01211789 AFYH01211790 AFYH01211791 AFYH01211792 AFYH01211793 AFYH01211794 AFYH01211795 AFYH01211796 AMQN01010558 KB308144 ELT98223.1 AC108840 AC123555 AC138680 DS469509 EDO49515.1 GBYX01227677 JAO69464.1 IACK01050379 LAA73508.1 JW867793 AFP00311.1 GBYX01227690 GBYX01227688 GBYX01227684 GBYX01227680 JAO69454.1 BEZZ01000995 GCC37407.1 AYCK01009418 IACK01050380 LAA73509.1 NCKU01003092 RWS08190.1 AKCR02000010 PKK29661.1 AHAT01011349 AHAT01011350 AHAT01011351 AHAT01011352 AHAT01011353 AHAT01011354 AHAT01011355 GCES01069483 JAR16840.1 GCES01069484 JAR16839.1 GCES01069482 JAR16841.1 LSYS01004331 OPJ80175.1 OPJ80174.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000079169
UP000007266
+ More
UP000030742 UP000019118 UP000009046 UP000015103 UP000030746 UP000245037 UP000265000 UP000288954 UP000242188 UP000001555 UP000261540 UP000005640 UP000008144 UP000076420 UP000007110 UP000265100 UP000245119 UP000245300 UP000002279 UP000005408 UP000008672 UP000192224 UP000014760 UP000000589 UP000085678 UP000007875 UP000001593 UP000087266 UP000261440 UP000287033 UP000261500 UP000261480 UP000002852 UP000028760 UP000242638 UP000192220 UP000248482 UP000285301 UP000053872 UP000018468 UP000190648 UP000264800
UP000030742 UP000019118 UP000009046 UP000015103 UP000030746 UP000245037 UP000265000 UP000288954 UP000242188 UP000001555 UP000261540 UP000005640 UP000008144 UP000076420 UP000007110 UP000265100 UP000245119 UP000245300 UP000002279 UP000005408 UP000008672 UP000192224 UP000014760 UP000000589 UP000085678 UP000007875 UP000001593 UP000087266 UP000261440 UP000287033 UP000261500 UP000261480 UP000002852 UP000028760 UP000242638 UP000192220 UP000248482 UP000285301 UP000053872 UP000018468 UP000190648 UP000264800
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A2W1BDI3
A0A2A4JWJ4
A0A2A4JUS6
A0A2A4JWI6
A0A2H1V8X2
A0A194Q2K8
+ More
A0A212ELY4 A0A0N1PI54 A0A1Y1K1E7 A0A3Q0IYS9 D6WBX6 U4UKC3 N6U416 A0A1B6HVB5 E0VD23 T1HXQ9 V4CKW9 A0A2P8XZ55 A0A1B6DN56 A0A0S7JXX8 A0A3Q2P298 A0A402G3X2 A0A210PRF8 A0A0B7BA75 A0A0B7B9V2 A0A0B7B8Z7 A0A0B7BCD9 B7Q0V0 A0A3B3T082 A0A3B3T034 C9JUP5 A0A2J8N4H2 A0A2J8WVQ8 C9JYW1 F6W622 A0A1W2WAC1 F6W6J2 A0A2C9MAE6 A0A2J8N4H0 A0A2J8WVS1 W4XUC0 A0A3P8P7F9 A0A2T7PV62 S4R706 F7G232 E7EQD9 K1RK57 Q05CR5 A0A2P4S4B9 H2ZWQ3 A0A1W4ZDJ7 R7TWD0 A0A1W4Z4U8 A0A1W4ZE34 A0A1W4ZDH8 A0A1W4Z4U7 A0A1W4ZDJ2 D3YV80 A0A1S3HRA3 A0A1S3HQK3 H2Z8V8 H2Z8V7 A7RGF6 A0A1S3PNB1 A0A0S7JW73 A0A2D4HNJ1 V9KQF7 A0A3B4CMM8 A0A3B4E642 A0A3B4E7L7 A0A3B4E6D9 A0A0S7JWJ0 A0A401T467 A0A3B3UL06 A0A3B3XM75 A0A3B5PU45 A0A087XEB4 A0A3B3UKB7 A0A3P9PM21 A0A3P9PM46 A0A3B5QR98 A0A3P9PM01 A0A3P9PLM7 A0A2I4BW27 A0A2D4HNF6 M4AQ39 A0A3P9PLF5 A0A2Y9L955 A0A2Y9L8Y0 A0A2Y9LED4 A0A3S3NY11 A0A2I0MJ07 W5MA88 A0A146VHI7 A0A146VIH2 A0A146VHH4 A0A1V4K708 A0A1V4K6Y0 A0A3Q3GGA1 A0A3Q3AZD4
A0A212ELY4 A0A0N1PI54 A0A1Y1K1E7 A0A3Q0IYS9 D6WBX6 U4UKC3 N6U416 A0A1B6HVB5 E0VD23 T1HXQ9 V4CKW9 A0A2P8XZ55 A0A1B6DN56 A0A0S7JXX8 A0A3Q2P298 A0A402G3X2 A0A210PRF8 A0A0B7BA75 A0A0B7B9V2 A0A0B7B8Z7 A0A0B7BCD9 B7Q0V0 A0A3B3T082 A0A3B3T034 C9JUP5 A0A2J8N4H2 A0A2J8WVQ8 C9JYW1 F6W622 A0A1W2WAC1 F6W6J2 A0A2C9MAE6 A0A2J8N4H0 A0A2J8WVS1 W4XUC0 A0A3P8P7F9 A0A2T7PV62 S4R706 F7G232 E7EQD9 K1RK57 Q05CR5 A0A2P4S4B9 H2ZWQ3 A0A1W4ZDJ7 R7TWD0 A0A1W4Z4U8 A0A1W4ZE34 A0A1W4ZDH8 A0A1W4Z4U7 A0A1W4ZDJ2 D3YV80 A0A1S3HRA3 A0A1S3HQK3 H2Z8V8 H2Z8V7 A7RGF6 A0A1S3PNB1 A0A0S7JW73 A0A2D4HNJ1 V9KQF7 A0A3B4CMM8 A0A3B4E642 A0A3B4E7L7 A0A3B4E6D9 A0A0S7JWJ0 A0A401T467 A0A3B3UL06 A0A3B3XM75 A0A3B5PU45 A0A087XEB4 A0A3B3UKB7 A0A3P9PM21 A0A3P9PM46 A0A3B5QR98 A0A3P9PM01 A0A3P9PLM7 A0A2I4BW27 A0A2D4HNF6 M4AQ39 A0A3P9PLF5 A0A2Y9L955 A0A2Y9L8Y0 A0A2Y9LED4 A0A3S3NY11 A0A2I0MJ07 W5MA88 A0A146VHI7 A0A146VIH2 A0A146VHH4 A0A1V4K708 A0A1V4K6Y0 A0A3Q3GGA1 A0A3Q3AZD4
PDB
6C1H
E-value=4.32737e-24,
Score=269
Ontologies
KEGG
GO
Topology
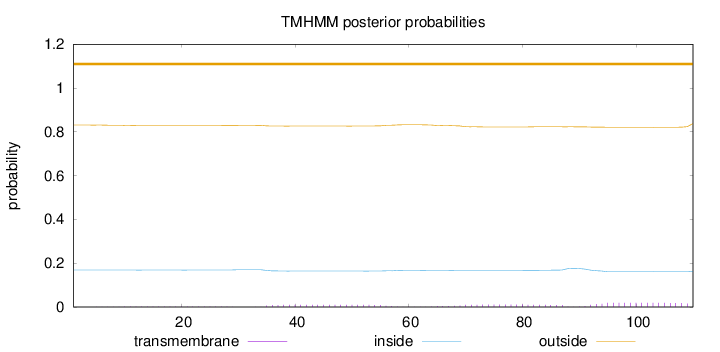
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.77613
Exp number, first 60 AAs:
0.25046
Total prob of N-in:
0.16894
outside
1 - 110
Population Genetic Test Statistics
Pi
12.903063
Theta
13.157173
Tajima's D
-0.419894
CLR
0.681006
CSRT
0.261786910654467
Interpretation
Uncertain
