Pre Gene Modal
BGIBMGA013462
Annotation
PREDICTED:_programmed_cell_death_6-interacting_protein_[Papilio_xuthus]
Full name
Histone H4
Location in the cell
Cytoplasmic Reliability : 2.533
Sequence
CDS
ATGGCTGAATTGTTGTTTGTGCCGTTTAAAAAGTCTTCGGACGTTGATATTGTTAAACCTTTGAAGAATTTAATACAATCTACTTACAACAGTGCCGAGAACACAGAAGATTATACCGATGCCTTAAATGAACTGAGCCGGTTGCGTACCAACGCAGTTTGGAAAGTCTTCGAGAAAACCTCTCTTGACACTATTTACAGTTACTATGATCAACTGGTGGCCCTTGAGAGCAAGATTCCTCCTCAAGAAGTTCAAATTCCATTCAAATGGAAGGATGCATTTGACAAGGGTTCTATCTTTGGAGGCAGGATGAGTCTCACAATCAGTTCATTGGAATATGAAAGAATGTGCATCTTATTCAACATAGCAGCTATGCAGAGCATGATTGCAGCTCAGGAACCTCTCGACACTGAGGATTCACTCAAGCTAGCTGCCAAATATTTTCAGCAATCAGCTGGCATCTTTGTATACCTGAAGGCGAACATCATGATGGCAGTGCACCAGGAAACCACACCAGATTTGAACCCTGAAACGCTGGATGCATTGGCCAAACTAATGCTCGCACAGGCACAGGAGGTTATCGCCCATAAATGCATCAGAGATGAGATGAAGGAGAGCATGGTGGCAAAGGTGTGCGCGCAGTGCGAGGAGCTGTACGTGGACGCGCTGCGGGCCATGCTCAAGGAGAACCCCAAGTCGCTCTGGGAAAAGGACTGGGTACCAAATGTTCAAGCGAAGCAGATGGCGTTCCGCGGCCTGGCGCAGTACTACCAGAGCCTGGTGTGCCGCGCCAACAAGGCCGTGGGCGAGGAGATCGCCAGACTACAATTCGCAGTAGATCAGCTGAAGGGTGCCCAGTCGCGGCTGTCGCCCAGCTACCTGCAGGAGCAGGCGTCGAAGGCAGCGCGCAACCTTCAAGAGGCGATGAAGGACAACGACTTCATCTACCACGAGCGCATCCCGGACCTCAAGCAGCTGGAGCTCATCTCCAAGGCGGCCATCGCTAAACCTTTGCCTCCACAAGAGAAGTGGGGCTCGGGGGGGAAAGATCTGTTCGCTGGGCTGGTCCCGCTGGCGGTGCACCACGCGGCGGCGGCCAGCAGCGCGCGCCGCGCCGAGCTGGTCTCCGCCGAGGTCGCCAAGCTGCGGGACGCCACGCAGCTGCTCAACAGCATCCTGGCGTCGCTGAACCTGCCGGCGTGCATCGAGTCGTGCGACGAGGAGGGCTCGCTGCCGGAGTCCATCCGCAGCAAGGCGCGCGCCGTGCGGGACGCGGGCGGCCTGCAAGAGCTGCAGCGCCTCATGGCCGAGCTGCCGGACCTGCTGCAGAGGAACAAGGACATCCTCGACGAGGCGGAGCGCATGCTGCGGGAGGAGGCGGAGGCGGACGCGGCGCTGCGGCAACAGTTCGGGGCCCGCTGGACGCGCACGCCCTCCGCCGCGCTCACCGAGGCCTTCCGCGCCAACGCGGACAAGTACCGGCAGATCATCGACAACGCCGTGCGCGCCGACAACATCGTGCAGCAGAAGTACCAGCAGCACAAGGACAACATAGCGCTGCTGAGCGGCAGCGAGGGCGACATCACGGCGGGGGTGCCCTCCGCGCCCGCCGCCGGGCCCGCCGACACGCGCGCACTCTCCACGCTGCGAGGTCTCATGGAGGAGGTGGAGAGGCTGAAGGCGGAGCGGGACAGCATCGAGTGCTCGCTGAAGGACGCGAGCGTGGACCTGCGGGAGCAGTTCCTGGCGGCGCTGAGCGCGGACGGCGCCATCGACGAGCCCGCGCTGTCCGCCGCCGCGCTGGGCCGCGCGCTGGGCCCGCTGCAGGCGCAGCTCGCCGCCTCGCTCGCCGCGCAGGACGACCTCGTGGCGCGCATACAGAAAGCCCACGGCAGCCTGGAGTCTGGCGGGGGGGGCGCGGGCGGGAGGGACGCGGTGCTGGGCCGCCTGGCCGCTGCCTACGACGCCTTCCAGGACCTCACCGGCAACCTCAGGGAGGGGGTCAAGTTCTACAACGACCTCACGCAGCTGCTGGTCGCCTTCCAAAACAAAGTATCGGACTTCTGTTTCGCTCGTAAGACTGAGAAGGAGGAGCTCCTCAAGGACCTCACCCAGGAGGCCTCCCGCCCCGCGACCAGGCCCGCCCCGAGCCCGCCGCAACACCACACCGACGCCGTGGCCGAGGTAAACAGAGTATTCTAA
Protein
MAELLFVPFKKSSDVDIVKPLKNLIQSTYNSAENTEDYTDALNELSRLRTNAVWKVFEKTSLDTIYSYYDQLVALESKIPPQEVQIPFKWKDAFDKGSIFGGRMSLTISSLEYERMCILFNIAAMQSMIAAQEPLDTEDSLKLAAKYFQQSAGIFVYLKANIMMAVHQETTPDLNPETLDALAKLMLAQAQEVIAHKCIRDEMKESMVAKVCAQCEELYVDALRAMLKENPKSLWEKDWVPNVQAKQMAFRGLAQYYQSLVCRANKAVGEEIARLQFAVDQLKGAQSRLSPSYLQEQASKAARNLQEAMKDNDFIYHERIPDLKQLELISKAAIAKPLPPQEKWGSGGKDLFAGLVPLAVHHAAAASSARRAELVSAEVAKLRDATQLLNSILASLNLPACIESCDEEGSLPESIRSKARAVRDAGGLQELQRLMAELPDLLQRNKDILDEAERMLREEAEADAALRQQFGARWTRTPSAALTEAFRANADKYRQIIDNAVRADNIVQQKYQQHKDNIALLSGSEGDITAGVPSAPAAGPADTRALSTLRGLMEEVERLKAERDSIECSLKDASVDLREQFLAALSADGAIDEPALSAAALGRALGPLQAQLAASLAAQDDLVARIQKAHGSLESGGGGAGGRDAVLGRLAAAYDAFQDLTGNLREGVKFYNDLTQLLVAFQNKVSDFCFARKTEKEELLKDLTQEASRPATRPAPSPPQHHTDAVAEVNRVF
Summary
Description
Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
Subunit
The nucleosome is a histone octamer containing two molecules each of H2A, H2B, H3 and H4 assembled in one H3-H4 heterotetramer and two H2A-H2B heterodimers. The octamer wraps approximately 147 bp of DNA.
Similarity
Belongs to the histone H4 family.
Uniprot
H9JV99
A0A2H1VQU3
A0A212ELZ9
A0A2J7PWQ8
A0A2J7PWR4
A0A437BHX4
+ More
A0A067RRI5 A0A1Y1LGS8 D6WD78 A0A154PAW4 A0A0L7RGL7 J3JTI1 A0A026WAB2 V5G0L5 A0A1B6HFU3 A0A151J4B4 E2BRJ3 A0A1I9WLI5 T1HX75 A0A0P4VM27 A0A088ANI4 A0A1B0GP78 V9IJ51 A0A1B6KHR9 A0A195FSG0 A0A069DX83 A0A1B6MPS5 A0A195BL44 A0A411G6S2 A0A1B6KH93 A0A0V0G4D7 A0A1L8DJF9 A0A023F397 A0A158NSG5 F4WFM8 A0A1B6EQG9 A0A0M8ZPZ9 A0A224XJK6 A0A1B6MA61 A0A1B6KGS5 A0A1B6MMN7 A5XB12 A0A0P4W5P3 T1JEE2 B4KAS5 A0A1W4VL42 A0A0M4F2Q1 A0A0A1X512 A0A3B0KAX2 B4JU60 T1PEF6 W8BYV1 A0A1B6MV22 B4M616 B4IGV4 B3LX31 B4QY34 A0A1I8P1A3 B4NAS1 B4PRA8 A0A1A9WC01 Q9VB05 A0A3B0K3Z2 Q29BG1 A0A1B0FP40 A0A1A9Z7A3 E0V9Q3 A0A034W514 B3P5Y2 A0A182HDP2 A0A444THJ6 A0A0K8V4B1 A0A023EVA8 A0A0P6ATJ7 A0A0P5APV9 A0A0N8A152 A0A0P5KN73 A0A0P6JF15 A0A0P6E396 A0A0L0BNC6 A0A2P2HW37 Q16QI6 A0A0P5TZM9 A0A0P4ZR37 A0A1A9ULX9 A0A2A3ECD3 A0A336M9W6 A0A1Q3FE56 A0A0A9XXL8 A0A146L7V4 A0A0P5PIU4 A0A0J7K5H8 T1E1D8 B4GNW9 E9G102 A0A084VZ78 A0A2R5LI09 A0A293MWL8 A0A2S2QYR3 A0A023GNS2
A0A067RRI5 A0A1Y1LGS8 D6WD78 A0A154PAW4 A0A0L7RGL7 J3JTI1 A0A026WAB2 V5G0L5 A0A1B6HFU3 A0A151J4B4 E2BRJ3 A0A1I9WLI5 T1HX75 A0A0P4VM27 A0A088ANI4 A0A1B0GP78 V9IJ51 A0A1B6KHR9 A0A195FSG0 A0A069DX83 A0A1B6MPS5 A0A195BL44 A0A411G6S2 A0A1B6KH93 A0A0V0G4D7 A0A1L8DJF9 A0A023F397 A0A158NSG5 F4WFM8 A0A1B6EQG9 A0A0M8ZPZ9 A0A224XJK6 A0A1B6MA61 A0A1B6KGS5 A0A1B6MMN7 A5XB12 A0A0P4W5P3 T1JEE2 B4KAS5 A0A1W4VL42 A0A0M4F2Q1 A0A0A1X512 A0A3B0KAX2 B4JU60 T1PEF6 W8BYV1 A0A1B6MV22 B4M616 B4IGV4 B3LX31 B4QY34 A0A1I8P1A3 B4NAS1 B4PRA8 A0A1A9WC01 Q9VB05 A0A3B0K3Z2 Q29BG1 A0A1B0FP40 A0A1A9Z7A3 E0V9Q3 A0A034W514 B3P5Y2 A0A182HDP2 A0A444THJ6 A0A0K8V4B1 A0A023EVA8 A0A0P6ATJ7 A0A0P5APV9 A0A0N8A152 A0A0P5KN73 A0A0P6JF15 A0A0P6E396 A0A0L0BNC6 A0A2P2HW37 Q16QI6 A0A0P5TZM9 A0A0P4ZR37 A0A1A9ULX9 A0A2A3ECD3 A0A336M9W6 A0A1Q3FE56 A0A0A9XXL8 A0A146L7V4 A0A0P5PIU4 A0A0J7K5H8 T1E1D8 B4GNW9 E9G102 A0A084VZ78 A0A2R5LI09 A0A293MWL8 A0A2S2QYR3 A0A023GNS2
Pubmed
19121390
22118469
24845553
28004739
18362917
19820115
+ More
22516182 23537049 24508170 30249741 20798317 27538518 27129103 26334808 25474469 21347285 21719571 17383199 17994087 25830018 25315136 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20566863 25348373 26483478 24945155 26108605 17510324 25401762 26823975 24330624 21292972 24438588
22516182 23537049 24508170 30249741 20798317 27538518 27129103 26334808 25474469 21347285 21719571 17383199 17994087 25830018 25315136 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20566863 25348373 26483478 24945155 26108605 17510324 25401762 26823975 24330624 21292972 24438588
EMBL
BABH01037335
BABH01037336
BABH01037337
BABH01037338
BABH01037339
ODYU01003886
+ More
SOQ43205.1 AGBW02013968 OWR42499.1 NEVH01020869 PNF20765.1 PNF20766.1 RSAL01000053 RVE50047.1 KK852470 KDR23230.1 GEZM01057943 GEZM01057942 JAV72081.1 KQ971312 EEZ98352.1 KQ434864 KZC08983.1 KQ414596 KOC69968.1 APGK01047880 APGK01047881 APGK01047882 BT126538 KB741092 KB632050 AEE61502.1 ENN73916.1 ERL88308.1 KK107295 QOIP01000003 EZA52990.1 RLU24287.1 GALX01004921 JAB63545.1 GECU01034184 JAS73522.1 KQ980159 KYN17416.1 GL449965 EFN81685.1 KU932369 APA34005.1 ACPB03011000 GDKW01000447 JAI56148.1 AJVK01014999 JR047269 AEY60369.1 GEBQ01028960 JAT11017.1 KQ981276 KYN43525.1 GBGD01000468 JAC88421.1 GEBQ01002098 JAT37879.1 KQ976444 KYM86247.1 MH365754 QBB01563.1 GEBQ01029161 JAT10816.1 GECL01003584 JAP02540.1 GFDF01007523 JAV06561.1 GBBI01003046 JAC15666.1 ADTU01024896 ADTU01024897 GL888120 EGI67005.1 GECZ01029570 JAS40199.1 KQ436008 KOX67616.1 GFTR01008113 JAW08313.1 GEBQ01007152 GEBQ01001159 JAT32825.1 JAT38818.1 GEBQ01029310 JAT10667.1 GEBQ01002771 JAT37206.1 DQ787160 ABG82044.1 GDRN01070718 JAI63819.1 JH432116 CH933806 EDW16812.1 CP012526 ALC45750.1 GBXI01007883 JAD06409.1 OUUW01000005 SPP80718.1 CH916374 EDV91030.1 KA647074 AFP61703.1 GAMC01002068 JAC04488.1 GEBQ01000214 JAT39763.1 CH940652 EDW59092.1 CH480837 EDW49072.1 CH902617 EDV43870.1 CM000364 EDX14682.1 CH964232 EDW80885.1 CM000160 EDW98472.1 AE014297 AY069534 AAF56740.1 AAL39679.1 SPP80719.1 CM000070 EAL27037.1 CCAG010023115 DS234995 EEB10088.1 GAKP01009550 JAC49402.1 CH954182 EDV53382.1 JXUM01130051 JXUM01130052 JXUM01130053 JXUM01130054 JXUM01130055 JXUM01130056 KQ567530 KXJ69388.1 SAUD01002650 RXG69456.1 GDHF01018889 JAI33425.1 GAPW01000213 JAC13385.1 GDIP01026445 JAM77270.1 GDIP01196175 JAJ27227.1 GDIP01192721 GDIP01090984 JAJ30681.1 GDIQ01189146 JAK62579.1 GDIQ01008511 JAN86226.1 GDIQ01190400 GDIQ01189147 GDIQ01068754 JAN25983.1 JRES01001623 KNC21423.1 IACF01000188 LAB65991.1 CH477747 EAT36654.1 GDIP01120278 JAL83436.1 GDIP01209968 JAJ13434.1 KZ288292 PBC29144.1 UFQT01000550 SSX25167.1 GFDL01009218 JAV25827.1 GBHO01019488 GBHO01010930 GBRD01012240 GBRD01012238 JAG24116.1 JAG32674.1 JAG53584.1 GDHC01014954 JAQ03675.1 GDIQ01149374 JAL02352.1 LBMM01013273 KMQ85728.1 GALA01001769 JAA93083.1 CH479186 EDW38852.1 GL732529 EFX86720.1 ATLV01018617 KE525239 KFB43272.1 GGLE01005025 MBY09151.1 GFWV01019613 MAA44341.1 GGMS01013059 MBY82262.1 GBBM01000540 JAC34878.1
SOQ43205.1 AGBW02013968 OWR42499.1 NEVH01020869 PNF20765.1 PNF20766.1 RSAL01000053 RVE50047.1 KK852470 KDR23230.1 GEZM01057943 GEZM01057942 JAV72081.1 KQ971312 EEZ98352.1 KQ434864 KZC08983.1 KQ414596 KOC69968.1 APGK01047880 APGK01047881 APGK01047882 BT126538 KB741092 KB632050 AEE61502.1 ENN73916.1 ERL88308.1 KK107295 QOIP01000003 EZA52990.1 RLU24287.1 GALX01004921 JAB63545.1 GECU01034184 JAS73522.1 KQ980159 KYN17416.1 GL449965 EFN81685.1 KU932369 APA34005.1 ACPB03011000 GDKW01000447 JAI56148.1 AJVK01014999 JR047269 AEY60369.1 GEBQ01028960 JAT11017.1 KQ981276 KYN43525.1 GBGD01000468 JAC88421.1 GEBQ01002098 JAT37879.1 KQ976444 KYM86247.1 MH365754 QBB01563.1 GEBQ01029161 JAT10816.1 GECL01003584 JAP02540.1 GFDF01007523 JAV06561.1 GBBI01003046 JAC15666.1 ADTU01024896 ADTU01024897 GL888120 EGI67005.1 GECZ01029570 JAS40199.1 KQ436008 KOX67616.1 GFTR01008113 JAW08313.1 GEBQ01007152 GEBQ01001159 JAT32825.1 JAT38818.1 GEBQ01029310 JAT10667.1 GEBQ01002771 JAT37206.1 DQ787160 ABG82044.1 GDRN01070718 JAI63819.1 JH432116 CH933806 EDW16812.1 CP012526 ALC45750.1 GBXI01007883 JAD06409.1 OUUW01000005 SPP80718.1 CH916374 EDV91030.1 KA647074 AFP61703.1 GAMC01002068 JAC04488.1 GEBQ01000214 JAT39763.1 CH940652 EDW59092.1 CH480837 EDW49072.1 CH902617 EDV43870.1 CM000364 EDX14682.1 CH964232 EDW80885.1 CM000160 EDW98472.1 AE014297 AY069534 AAF56740.1 AAL39679.1 SPP80719.1 CM000070 EAL27037.1 CCAG010023115 DS234995 EEB10088.1 GAKP01009550 JAC49402.1 CH954182 EDV53382.1 JXUM01130051 JXUM01130052 JXUM01130053 JXUM01130054 JXUM01130055 JXUM01130056 KQ567530 KXJ69388.1 SAUD01002650 RXG69456.1 GDHF01018889 JAI33425.1 GAPW01000213 JAC13385.1 GDIP01026445 JAM77270.1 GDIP01196175 JAJ27227.1 GDIP01192721 GDIP01090984 JAJ30681.1 GDIQ01189146 JAK62579.1 GDIQ01008511 JAN86226.1 GDIQ01190400 GDIQ01189147 GDIQ01068754 JAN25983.1 JRES01001623 KNC21423.1 IACF01000188 LAB65991.1 CH477747 EAT36654.1 GDIP01120278 JAL83436.1 GDIP01209968 JAJ13434.1 KZ288292 PBC29144.1 UFQT01000550 SSX25167.1 GFDL01009218 JAV25827.1 GBHO01019488 GBHO01010930 GBRD01012240 GBRD01012238 JAG24116.1 JAG32674.1 JAG53584.1 GDHC01014954 JAQ03675.1 GDIQ01149374 JAL02352.1 LBMM01013273 KMQ85728.1 GALA01001769 JAA93083.1 CH479186 EDW38852.1 GL732529 EFX86720.1 ATLV01018617 KE525239 KFB43272.1 GGLE01005025 MBY09151.1 GFWV01019613 MAA44341.1 GGMS01013059 MBY82262.1 GBBM01000540 JAC34878.1
Proteomes
UP000005204
UP000007151
UP000235965
UP000283053
UP000027135
UP000007266
+ More
UP000076502 UP000053825 UP000019118 UP000030742 UP000053097 UP000279307 UP000078492 UP000008237 UP000015103 UP000005203 UP000092462 UP000078541 UP000078540 UP000005205 UP000007755 UP000053105 UP000009192 UP000192221 UP000092553 UP000268350 UP000001070 UP000095301 UP000008792 UP000001292 UP000007801 UP000000304 UP000095300 UP000007798 UP000002282 UP000091820 UP000000803 UP000001819 UP000092444 UP000092445 UP000009046 UP000008711 UP000069940 UP000249989 UP000288706 UP000037069 UP000008820 UP000078200 UP000242457 UP000036403 UP000008744 UP000000305 UP000030765
UP000076502 UP000053825 UP000019118 UP000030742 UP000053097 UP000279307 UP000078492 UP000008237 UP000015103 UP000005203 UP000092462 UP000078541 UP000078540 UP000005205 UP000007755 UP000053105 UP000009192 UP000192221 UP000092553 UP000268350 UP000001070 UP000095301 UP000008792 UP000001292 UP000007801 UP000000304 UP000095300 UP000007798 UP000002282 UP000091820 UP000000803 UP000001819 UP000092444 UP000092445 UP000009046 UP000008711 UP000069940 UP000249989 UP000288706 UP000037069 UP000008820 UP000078200 UP000242457 UP000036403 UP000008744 UP000000305 UP000030765
Pfam
Interpro
IPR004328
BRO1_dom
+ More
IPR038499 BRO1_sf
IPR025304 ALIX_V_dom
IPR019809 Histone_H4_CS
IPR001951 Histone_H4
IPR004823 TAF_TATA-bd
IPR035425 CENP-T/H4_C
IPR009072 Histone-fold
IPR022967 S1_dom
IPR007502 Helicase-assoc_dom
IPR003029 S1_domain
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR011709 DUF1605
IPR012340 NA-bd_OB-fold
IPR038499 BRO1_sf
IPR025304 ALIX_V_dom
IPR019809 Histone_H4_CS
IPR001951 Histone_H4
IPR004823 TAF_TATA-bd
IPR035425 CENP-T/H4_C
IPR009072 Histone-fold
IPR022967 S1_dom
IPR007502 Helicase-assoc_dom
IPR003029 S1_domain
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR011709 DUF1605
IPR012340 NA-bd_OB-fold
Gene 3D
ProteinModelPortal
H9JV99
A0A2H1VQU3
A0A212ELZ9
A0A2J7PWQ8
A0A2J7PWR4
A0A437BHX4
+ More
A0A067RRI5 A0A1Y1LGS8 D6WD78 A0A154PAW4 A0A0L7RGL7 J3JTI1 A0A026WAB2 V5G0L5 A0A1B6HFU3 A0A151J4B4 E2BRJ3 A0A1I9WLI5 T1HX75 A0A0P4VM27 A0A088ANI4 A0A1B0GP78 V9IJ51 A0A1B6KHR9 A0A195FSG0 A0A069DX83 A0A1B6MPS5 A0A195BL44 A0A411G6S2 A0A1B6KH93 A0A0V0G4D7 A0A1L8DJF9 A0A023F397 A0A158NSG5 F4WFM8 A0A1B6EQG9 A0A0M8ZPZ9 A0A224XJK6 A0A1B6MA61 A0A1B6KGS5 A0A1B6MMN7 A5XB12 A0A0P4W5P3 T1JEE2 B4KAS5 A0A1W4VL42 A0A0M4F2Q1 A0A0A1X512 A0A3B0KAX2 B4JU60 T1PEF6 W8BYV1 A0A1B6MV22 B4M616 B4IGV4 B3LX31 B4QY34 A0A1I8P1A3 B4NAS1 B4PRA8 A0A1A9WC01 Q9VB05 A0A3B0K3Z2 Q29BG1 A0A1B0FP40 A0A1A9Z7A3 E0V9Q3 A0A034W514 B3P5Y2 A0A182HDP2 A0A444THJ6 A0A0K8V4B1 A0A023EVA8 A0A0P6ATJ7 A0A0P5APV9 A0A0N8A152 A0A0P5KN73 A0A0P6JF15 A0A0P6E396 A0A0L0BNC6 A0A2P2HW37 Q16QI6 A0A0P5TZM9 A0A0P4ZR37 A0A1A9ULX9 A0A2A3ECD3 A0A336M9W6 A0A1Q3FE56 A0A0A9XXL8 A0A146L7V4 A0A0P5PIU4 A0A0J7K5H8 T1E1D8 B4GNW9 E9G102 A0A084VZ78 A0A2R5LI09 A0A293MWL8 A0A2S2QYR3 A0A023GNS2
A0A067RRI5 A0A1Y1LGS8 D6WD78 A0A154PAW4 A0A0L7RGL7 J3JTI1 A0A026WAB2 V5G0L5 A0A1B6HFU3 A0A151J4B4 E2BRJ3 A0A1I9WLI5 T1HX75 A0A0P4VM27 A0A088ANI4 A0A1B0GP78 V9IJ51 A0A1B6KHR9 A0A195FSG0 A0A069DX83 A0A1B6MPS5 A0A195BL44 A0A411G6S2 A0A1B6KH93 A0A0V0G4D7 A0A1L8DJF9 A0A023F397 A0A158NSG5 F4WFM8 A0A1B6EQG9 A0A0M8ZPZ9 A0A224XJK6 A0A1B6MA61 A0A1B6KGS5 A0A1B6MMN7 A5XB12 A0A0P4W5P3 T1JEE2 B4KAS5 A0A1W4VL42 A0A0M4F2Q1 A0A0A1X512 A0A3B0KAX2 B4JU60 T1PEF6 W8BYV1 A0A1B6MV22 B4M616 B4IGV4 B3LX31 B4QY34 A0A1I8P1A3 B4NAS1 B4PRA8 A0A1A9WC01 Q9VB05 A0A3B0K3Z2 Q29BG1 A0A1B0FP40 A0A1A9Z7A3 E0V9Q3 A0A034W514 B3P5Y2 A0A182HDP2 A0A444THJ6 A0A0K8V4B1 A0A023EVA8 A0A0P6ATJ7 A0A0P5APV9 A0A0N8A152 A0A0P5KN73 A0A0P6JF15 A0A0P6E396 A0A0L0BNC6 A0A2P2HW37 Q16QI6 A0A0P5TZM9 A0A0P4ZR37 A0A1A9ULX9 A0A2A3ECD3 A0A336M9W6 A0A1Q3FE56 A0A0A9XXL8 A0A146L7V4 A0A0P5PIU4 A0A0J7K5H8 T1E1D8 B4GNW9 E9G102 A0A084VZ78 A0A2R5LI09 A0A293MWL8 A0A2S2QYR3 A0A023GNS2
PDB
2OEV
E-value=7.56021e-119,
Score=1096
Ontologies
GO
GO:0003677
GO:0000786
GO:0005634
GO:0046982
GO:0006352
GO:0048133
GO:0005813
GO:0030041
GO:0000281
GO:0070938
GO:0048132
GO:0045171
GO:0046330
GO:1990182
GO:0030496
GO:0043162
GO:0005768
GO:0003676
GO:0005524
GO:0004386
GO:0016021
GO:0005515
GO:0003723
GO:0008270
GO:0005615
GO:0045211
GO:0006281
GO:0048384
Topology
Subcellular location
Chromosome
Nucleus
Nucleus
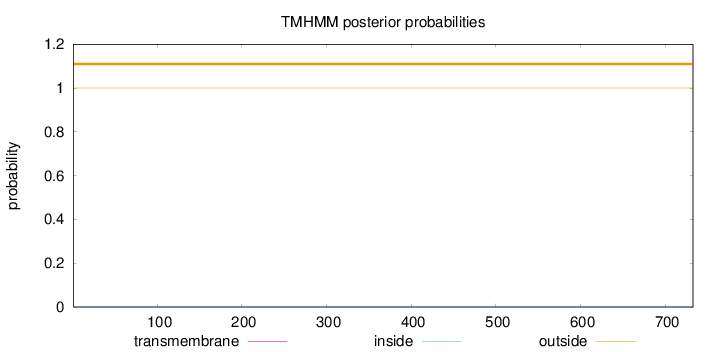
Length:
733
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00109
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00006
outside
1 - 733
Population Genetic Test Statistics
Pi
320.577449
Theta
183.941128
Tajima's D
2.419688
CLR
0.431736
CSRT
0.935653217339133
Interpretation
Uncertain
