Gene
KWMTBOMO03175
Pre Gene Modal
BGIBMGA013463
Annotation
PREDICTED:_MOB_kinase_activator-like_1_[Papilio_polytes]
Full name
MOB kinase activator-like 1
Alternative Name
Mob as tumor suppressor protein 1
Mps one binder kinase activator-like 1
Mps one binder kinase activator-like 1
Location in the cell
Cytoplasmic Reliability : 2.001
Sequence
CDS
ATGAGCTTCCTATTTGGTAGTCGCTCAAACAAAACTTTCAAACCAAAGAAGAATATTCCTGAGGGCACCCATCAGTACGATTTGATGAGGCACGCAGCAGCCACACTTGGTTCCGGGAACTTGCGCCTCGCTGTTATGTTACCCGAAGGGGAGGACCTCAATGAGTGGGTTGCCGTGAACACTGTGGATTTCTTCAATCAGATCAACATGTTATATGGGACCATAACAGAATTCTGCACAGAGGAGTCATGTGCTGTCATGTCTGCCGGACCAAAATATGAATACCATTGGGCTGACGGACACACAGTGAAGAAACCTATCAAGTGTTCGGCCCCAAAGTATATAGATTATTTGATGACGTGGGCACAGGATCAGCTTGATGACGAAACCCTGTTCCCTTCGAAAATTGGTGTGCCATTCCCGAAGAGTTTCCTACTGATGGCAAAAACTATATTGAAGCGTTTGTTCCGCGTGTACGCGCACATCTACCACCAGCACTTCCCGGAGGTGGTGCAGCTGGGGGAAGAGGCTCACCTCAATACGTCATTCAAACATTTTATATTCTTTGTACAGGAGTTTAATCTGATCGAGCGCAAAGAGTTAGCGCCCTTACAGGAGCTCATCGACAAACTGACAGCGAAGGACGCGCGATGA
Protein
MSFLFGSRSNKTFKPKKNIPEGTHQYDLMRHAAATLGSGNLRLAVMLPEGEDLNEWVAVNTVDFFNQINMLYGTITEFCTEESCAVMSAGPKYEYHWADGHTVKKPIKCSAPKYIDYLMTWAQDQLDDETLFPSKIGVPFPKSFLLMAKTILKRLFRVYAHIYHQHFPEVVQLGEEAHLNTSFKHFIFFVQEFNLIERKELAPLQELIDKLTAKDAR
Summary
Description
Coactivator of Warts (Wts) kinase in the Hippo/SWH (Sav/Wts/Hpo)signaling pathway, a signaling pathway that plays a pivotal role in organ size control and tumor suppression by restricting proliferation and promoting apoptosis. The core of this pathway is composed of a kinase cascade wherein Hippo (Hpo), in complex with its regulatory protein Salvador (Sav), phosphorylates and activates Warts (Wts) in complex with its regulatory protein Mats, which in turn phosphorylates and inactivates the Yorkie (Yki)oncoprotein. The Hippo/SWH signaling pathway inhibits the activity of the transcriptional complex formed by Scalloped (sd) and Yki and the target genes of this pathway include cyclin-E (cycE), diap1 and bantam. Mats is essential for early development and is required for proper chromosomal segregation in developing embryos.
Subunit
Interacts with and activates trc and wts.
Similarity
Belongs to the MOB1/phocein family.
Keywords
Apoptosis
Cell membrane
Chromosome partition
Complete proteome
Cytoplasm
Cytoskeleton
Developmental protein
Membrane
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Zinc
Feature
chain MOB kinase activator-like 1
Uniprot
H9JVA0
A0A212ELZ8
A0A2A4J2U2
A0A3S2NFW3
A0A2H1VAB1
A0A1E1WLE1
+ More
A0A2A3E2E4 V9ILH2 A0A088ABP9 A0A158NT28 A0A310SFS1 A0A2W1BHZ1 E2C8F5 A0A1B6DB19 A0A0T6ATU7 A0A1B6LYH4 A0A1B6JXF1 A0A1B6F1B5 A0A0L7QQS7 A0A1Y1L947 A0A1W4WW80 T1IPE9 A0A1S4ELC1 X5J6H4 D6X544 A0A2R7WNU9 J3JY93 A0A154PFT6 A0A195BHG8 A0A2J7QI29 K7J160 A0A0A9VRJ9 A0A0A9VT23 E9IBE6 A0A3L8D714 A6YPT6 A0A069DQP2 A0A232FI84 B4NH61 B4K5C9 A0A1B0CDJ9 A0A1S3JE19 A0A0L0CJJ7 B4G4N9 B4JTX7 A0A151XBS8 A0A3B0KVZ4 B5DYN3 B4LXU3 A0A2T7PZP6 A0A1L8DKX1 B3M227 A0A1D2MM14 A0A1I8MFM9 A0A2P6L9A8 A0A1A9XMA6 A0A1B0BWG5 W8BMG8 A0A0K8U5Q1 A0A034WQX3 A0A1W4W2L5 E9GD71 A0A0P5NX87 A0A1A9V8R9 A0A1B0A7V2 A0A1A9WNM0 D3TRA2 A0A1I8PJ80 A0A2R5LNB3 V4AN84 A0A067R962 A0A2P2I9U2 A0A131Y244 A0A0B6YXY1 A0A195F0K0 A0A087TGV5 A0A3S3P803 B4R014 B4PLP8 B4HE84 A0A0B4KGK3 Q95RA8 F4WG46 B3P7J0 A0A0P4WG65 C1BQJ6 A0A0K8RHM4 G3MSP7 A0A224Z0X5 A0A131YX18 L7M4F8 A0A131XKK8 A7SEP4 T1GFB1 A0A0K8TT26 A0A293MGX9 D3PFP5 A0A0L8HMV0 A0A336M2E2 A0A3S1BPR3
A0A2A3E2E4 V9ILH2 A0A088ABP9 A0A158NT28 A0A310SFS1 A0A2W1BHZ1 E2C8F5 A0A1B6DB19 A0A0T6ATU7 A0A1B6LYH4 A0A1B6JXF1 A0A1B6F1B5 A0A0L7QQS7 A0A1Y1L947 A0A1W4WW80 T1IPE9 A0A1S4ELC1 X5J6H4 D6X544 A0A2R7WNU9 J3JY93 A0A154PFT6 A0A195BHG8 A0A2J7QI29 K7J160 A0A0A9VRJ9 A0A0A9VT23 E9IBE6 A0A3L8D714 A6YPT6 A0A069DQP2 A0A232FI84 B4NH61 B4K5C9 A0A1B0CDJ9 A0A1S3JE19 A0A0L0CJJ7 B4G4N9 B4JTX7 A0A151XBS8 A0A3B0KVZ4 B5DYN3 B4LXU3 A0A2T7PZP6 A0A1L8DKX1 B3M227 A0A1D2MM14 A0A1I8MFM9 A0A2P6L9A8 A0A1A9XMA6 A0A1B0BWG5 W8BMG8 A0A0K8U5Q1 A0A034WQX3 A0A1W4W2L5 E9GD71 A0A0P5NX87 A0A1A9V8R9 A0A1B0A7V2 A0A1A9WNM0 D3TRA2 A0A1I8PJ80 A0A2R5LNB3 V4AN84 A0A067R962 A0A2P2I9U2 A0A131Y244 A0A0B6YXY1 A0A195F0K0 A0A087TGV5 A0A3S3P803 B4R014 B4PLP8 B4HE84 A0A0B4KGK3 Q95RA8 F4WG46 B3P7J0 A0A0P4WG65 C1BQJ6 A0A0K8RHM4 G3MSP7 A0A224Z0X5 A0A131YX18 L7M4F8 A0A131XKK8 A7SEP4 T1GFB1 A0A0K8TT26 A0A293MGX9 D3PFP5 A0A0L8HMV0 A0A336M2E2 A0A3S1BPR3
Pubmed
19121390
22118469
21347285
28756777
20798317
28004739
+ More
18362917 19820115 22516182 20075255 25401762 26823975 21282665 30249741 18207082 26334808 28648823 17994087 26108605 15632085 27289101 25315136 24495485 25348373 21292972 20353571 23254933 24845553 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 15766530 15975907 17347649 18245354 19913529 21719571 22216098 28797301 26830274 25576852 28049606 17615350 26369729
18362917 19820115 22516182 20075255 25401762 26823975 21282665 30249741 18207082 26334808 28648823 17994087 26108605 15632085 27289101 25315136 24495485 25348373 21292972 20353571 23254933 24845553 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 15766530 15975907 17347649 18245354 19913529 21719571 22216098 28797301 26830274 25576852 28049606 17615350 26369729
EMBL
BABH01037339
AGBW02013968
OWR42497.1
NWSH01003364
PCG66467.1
RSAL01000053
+ More
RVE50045.1 ODYU01001478 SOQ37726.1 GDQN01003286 JAT87768.1 KZ288466 PBC25436.1 JR051585 AEY61522.1 ADTU01025480 KQ765605 OAD53876.1 KZ150187 PZC72446.1 GL453666 EFN75790.1 GEDC01028482 GEDC01014415 JAS08816.1 JAS22883.1 LJIG01022821 KRT78545.1 GEBQ01011229 JAT28748.1 GECU01003854 JAT03853.1 GECZ01029713 GECZ01028659 GECZ01025727 JAS40056.1 JAS41110.1 JAS44042.1 KQ414786 KOC60846.1 GEZM01066976 JAV67607.1 JH431260 HF969261 CCX34990.1 KQ971382 EEZ97199.1 KK855177 PTY21292.1 BT128217 AEE63178.1 KQ434896 KZC10719.1 KQ976488 KYM83611.1 NEVH01013964 PNF28229.1 GBHO01045217 GBHO01045215 GBRD01002085 GDHC01000451 JAF98386.1 JAF98388.1 JAG63736.1 JAQ18178.1 GBHO01045218 JAF98385.1 GL762111 EFZ22023.1 QOIP01000012 RLU16011.1 EF639097 ABR27982.1 GBGD01002868 JAC86021.1 NNAY01000180 OXU30240.1 CH964272 EDW84558.1 CH933806 EDW16155.2 AJWK01007944 JRES01000310 KNC32420.1 CH479179 EDW24555.1 CH916374 EDV91556.1 KQ982320 KYQ57814.1 OUUW01000013 SPP88138.1 CM000070 EDY68014.2 CH940650 EDW67902.1 PZQS01000001 PVD38870.1 GFDF01007023 JAV07061.1 CH902617 EDV43351.1 LJIJ01000890 ODM93915.1 MWRG01000847 PRD35168.1 JXJN01021768 GAMC01006588 JAB99967.1 GDHF01030396 JAI21918.1 GAKP01002357 JAC56595.1 GL732539 EFX82745.1 GDIQ01136452 LRGB01000944 JAL15274.1 KZS14646.1 CCAG010015572 EZ423954 ADD20230.1 GGLE01006853 MBY10979.1 KB201234 ESO98617.1 KK852854 KDR15007.1 IACF01005205 LAB70791.1 GEFM01002477 JAP73319.1 HACG01014364 CEK61229.1 KQ981897 KYN33682.1 KK115167 KFM64344.1 NCKU01003595 NCKU01003589 RWS07230.1 RWS07244.1 CM000364 EDX13920.1 CM000160 EDW97997.1 CH480815 EDW43181.1 AE014297 AGB96215.1 AHN57464.1 AY061520 AAF55993.2 AAL29068.1 GL888128 EGI66813.1 CH954182 EDV54151.1 GDRN01050809 JAI66442.1 BT076875 ACO11299.1 GADI01003789 JAA70019.1 JO844898 AEO36515.1 GFPF01008524 MAA19670.1 GEDV01004824 JAP83733.1 GACK01005833 JAA59201.1 GEFH01000547 JAP68034.1 DS469638 EDO37824.1 CAQQ02126288 GDAI01000312 JAI17291.1 GFWV01015425 MAA40154.1 BT120451 ADD24091.1 KQ417740 KOF90532.1 UFQT01000109 SSX20128.1 RQTK01000081 RUS88512.1
RVE50045.1 ODYU01001478 SOQ37726.1 GDQN01003286 JAT87768.1 KZ288466 PBC25436.1 JR051585 AEY61522.1 ADTU01025480 KQ765605 OAD53876.1 KZ150187 PZC72446.1 GL453666 EFN75790.1 GEDC01028482 GEDC01014415 JAS08816.1 JAS22883.1 LJIG01022821 KRT78545.1 GEBQ01011229 JAT28748.1 GECU01003854 JAT03853.1 GECZ01029713 GECZ01028659 GECZ01025727 JAS40056.1 JAS41110.1 JAS44042.1 KQ414786 KOC60846.1 GEZM01066976 JAV67607.1 JH431260 HF969261 CCX34990.1 KQ971382 EEZ97199.1 KK855177 PTY21292.1 BT128217 AEE63178.1 KQ434896 KZC10719.1 KQ976488 KYM83611.1 NEVH01013964 PNF28229.1 GBHO01045217 GBHO01045215 GBRD01002085 GDHC01000451 JAF98386.1 JAF98388.1 JAG63736.1 JAQ18178.1 GBHO01045218 JAF98385.1 GL762111 EFZ22023.1 QOIP01000012 RLU16011.1 EF639097 ABR27982.1 GBGD01002868 JAC86021.1 NNAY01000180 OXU30240.1 CH964272 EDW84558.1 CH933806 EDW16155.2 AJWK01007944 JRES01000310 KNC32420.1 CH479179 EDW24555.1 CH916374 EDV91556.1 KQ982320 KYQ57814.1 OUUW01000013 SPP88138.1 CM000070 EDY68014.2 CH940650 EDW67902.1 PZQS01000001 PVD38870.1 GFDF01007023 JAV07061.1 CH902617 EDV43351.1 LJIJ01000890 ODM93915.1 MWRG01000847 PRD35168.1 JXJN01021768 GAMC01006588 JAB99967.1 GDHF01030396 JAI21918.1 GAKP01002357 JAC56595.1 GL732539 EFX82745.1 GDIQ01136452 LRGB01000944 JAL15274.1 KZS14646.1 CCAG010015572 EZ423954 ADD20230.1 GGLE01006853 MBY10979.1 KB201234 ESO98617.1 KK852854 KDR15007.1 IACF01005205 LAB70791.1 GEFM01002477 JAP73319.1 HACG01014364 CEK61229.1 KQ981897 KYN33682.1 KK115167 KFM64344.1 NCKU01003595 NCKU01003589 RWS07230.1 RWS07244.1 CM000364 EDX13920.1 CM000160 EDW97997.1 CH480815 EDW43181.1 AE014297 AGB96215.1 AHN57464.1 AY061520 AAF55993.2 AAL29068.1 GL888128 EGI66813.1 CH954182 EDV54151.1 GDRN01050809 JAI66442.1 BT076875 ACO11299.1 GADI01003789 JAA70019.1 JO844898 AEO36515.1 GFPF01008524 MAA19670.1 GEDV01004824 JAP83733.1 GACK01005833 JAA59201.1 GEFH01000547 JAP68034.1 DS469638 EDO37824.1 CAQQ02126288 GDAI01000312 JAI17291.1 GFWV01015425 MAA40154.1 BT120451 ADD24091.1 KQ417740 KOF90532.1 UFQT01000109 SSX20128.1 RQTK01000081 RUS88512.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000242457
UP000005203
+ More
UP000005205 UP000008237 UP000053825 UP000192223 UP000079169 UP000007266 UP000076502 UP000078540 UP000235965 UP000002358 UP000279307 UP000215335 UP000007798 UP000009192 UP000092461 UP000085678 UP000037069 UP000008744 UP000001070 UP000075809 UP000268350 UP000001819 UP000008792 UP000245119 UP000007801 UP000094527 UP000095301 UP000092443 UP000092460 UP000192221 UP000000305 UP000076858 UP000078200 UP000092445 UP000091820 UP000092444 UP000095300 UP000030746 UP000027135 UP000078541 UP000054359 UP000285301 UP000000304 UP000002282 UP000001292 UP000000803 UP000007755 UP000008711 UP000001593 UP000015102 UP000053454 UP000271974
UP000005205 UP000008237 UP000053825 UP000192223 UP000079169 UP000007266 UP000076502 UP000078540 UP000235965 UP000002358 UP000279307 UP000215335 UP000007798 UP000009192 UP000092461 UP000085678 UP000037069 UP000008744 UP000001070 UP000075809 UP000268350 UP000001819 UP000008792 UP000245119 UP000007801 UP000094527 UP000095301 UP000092443 UP000092460 UP000192221 UP000000305 UP000076858 UP000078200 UP000092445 UP000091820 UP000092444 UP000095300 UP000030746 UP000027135 UP000078541 UP000054359 UP000285301 UP000000304 UP000002282 UP000001292 UP000000803 UP000007755 UP000008711 UP000001593 UP000015102 UP000053454 UP000271974
Pfam
PF03637 Mob1_phocein
SUPFAM
SSF101152
SSF101152
Gene 3D
ProteinModelPortal
H9JVA0
A0A212ELZ8
A0A2A4J2U2
A0A3S2NFW3
A0A2H1VAB1
A0A1E1WLE1
+ More
A0A2A3E2E4 V9ILH2 A0A088ABP9 A0A158NT28 A0A310SFS1 A0A2W1BHZ1 E2C8F5 A0A1B6DB19 A0A0T6ATU7 A0A1B6LYH4 A0A1B6JXF1 A0A1B6F1B5 A0A0L7QQS7 A0A1Y1L947 A0A1W4WW80 T1IPE9 A0A1S4ELC1 X5J6H4 D6X544 A0A2R7WNU9 J3JY93 A0A154PFT6 A0A195BHG8 A0A2J7QI29 K7J160 A0A0A9VRJ9 A0A0A9VT23 E9IBE6 A0A3L8D714 A6YPT6 A0A069DQP2 A0A232FI84 B4NH61 B4K5C9 A0A1B0CDJ9 A0A1S3JE19 A0A0L0CJJ7 B4G4N9 B4JTX7 A0A151XBS8 A0A3B0KVZ4 B5DYN3 B4LXU3 A0A2T7PZP6 A0A1L8DKX1 B3M227 A0A1D2MM14 A0A1I8MFM9 A0A2P6L9A8 A0A1A9XMA6 A0A1B0BWG5 W8BMG8 A0A0K8U5Q1 A0A034WQX3 A0A1W4W2L5 E9GD71 A0A0P5NX87 A0A1A9V8R9 A0A1B0A7V2 A0A1A9WNM0 D3TRA2 A0A1I8PJ80 A0A2R5LNB3 V4AN84 A0A067R962 A0A2P2I9U2 A0A131Y244 A0A0B6YXY1 A0A195F0K0 A0A087TGV5 A0A3S3P803 B4R014 B4PLP8 B4HE84 A0A0B4KGK3 Q95RA8 F4WG46 B3P7J0 A0A0P4WG65 C1BQJ6 A0A0K8RHM4 G3MSP7 A0A224Z0X5 A0A131YX18 L7M4F8 A0A131XKK8 A7SEP4 T1GFB1 A0A0K8TT26 A0A293MGX9 D3PFP5 A0A0L8HMV0 A0A336M2E2 A0A3S1BPR3
A0A2A3E2E4 V9ILH2 A0A088ABP9 A0A158NT28 A0A310SFS1 A0A2W1BHZ1 E2C8F5 A0A1B6DB19 A0A0T6ATU7 A0A1B6LYH4 A0A1B6JXF1 A0A1B6F1B5 A0A0L7QQS7 A0A1Y1L947 A0A1W4WW80 T1IPE9 A0A1S4ELC1 X5J6H4 D6X544 A0A2R7WNU9 J3JY93 A0A154PFT6 A0A195BHG8 A0A2J7QI29 K7J160 A0A0A9VRJ9 A0A0A9VT23 E9IBE6 A0A3L8D714 A6YPT6 A0A069DQP2 A0A232FI84 B4NH61 B4K5C9 A0A1B0CDJ9 A0A1S3JE19 A0A0L0CJJ7 B4G4N9 B4JTX7 A0A151XBS8 A0A3B0KVZ4 B5DYN3 B4LXU3 A0A2T7PZP6 A0A1L8DKX1 B3M227 A0A1D2MM14 A0A1I8MFM9 A0A2P6L9A8 A0A1A9XMA6 A0A1B0BWG5 W8BMG8 A0A0K8U5Q1 A0A034WQX3 A0A1W4W2L5 E9GD71 A0A0P5NX87 A0A1A9V8R9 A0A1B0A7V2 A0A1A9WNM0 D3TRA2 A0A1I8PJ80 A0A2R5LNB3 V4AN84 A0A067R962 A0A2P2I9U2 A0A131Y244 A0A0B6YXY1 A0A195F0K0 A0A087TGV5 A0A3S3P803 B4R014 B4PLP8 B4HE84 A0A0B4KGK3 Q95RA8 F4WG46 B3P7J0 A0A0P4WG65 C1BQJ6 A0A0K8RHM4 G3MSP7 A0A224Z0X5 A0A131YX18 L7M4F8 A0A131XKK8 A7SEP4 T1GFB1 A0A0K8TT26 A0A293MGX9 D3PFP5 A0A0L8HMV0 A0A336M2E2 A0A3S1BPR3
PDB
5B5V
E-value=2.4501e-113,
Score=1042
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Microtubule organizing center
Centrosome
Nucleus
Cytosol
Cell membrane
Cytoskeleton
Microtubule organizing center
Centrosome
Nucleus
Cytosol
Cell membrane
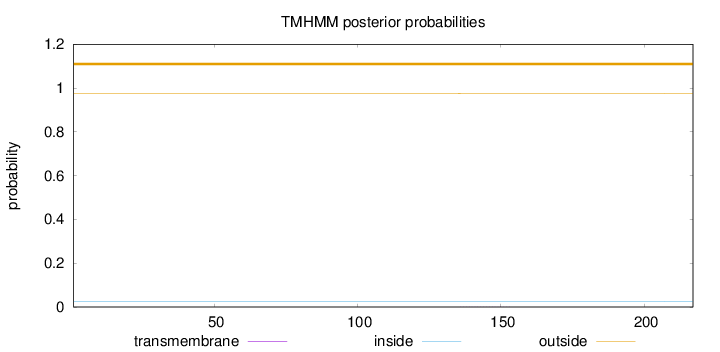
Length:
217
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00514
Exp number, first 60 AAs:
0.00029
Total prob of N-in:
0.02513
outside
1 - 217
Population Genetic Test Statistics
Pi
62.288061
Theta
145.585075
Tajima's D
-0.453204
CLR
2.064247
CSRT
0.250287485625719
Interpretation
Uncertain
