Gene
KWMTBOMO03168
Pre Gene Modal
BGIBMGA013446
Annotation
PREDICTED:_uncharacterized_protein_LOC106134809_[Amyelois_transitella]
Full name
MICOS complex subunit MIC60
Alternative Name
Mitofilin
Location in the cell
Cytoplasmic Reliability : 1.275 Mitochondrial Reliability : 1.012 Nuclear Reliability : 1.401
Sequence
CDS
ATGGTCCGCGATGGGATACGTCCCTTTGACTTCCTTCGTCAGTTCGATCCTCGTAACGAGCTGGTGATTCCTCGGGCCGACTTCTACCGAGGACTGGCTTCTGCAGGACTTTCACTGACGCCACTCGAAATGGATACGCTAATGGAAGTTTTCAGCGCCCCGGGTCGACGTAGATACGTGGAGTACGCTAGGTTCTCGTCCACCGTGGGCGAGGCTCTCACACAAGAAGGCCTAGAACGCGCACCGCTGCTGGTGCCAGTACCCCACCTCCCCACAGTAGACACGCCCCTCAACTACCTGAACTTCGAGGAACGGAATATTGTGTCCGGAGCTCTTGCCAAACTTGCAAAATATCCTGATCAGTTGTCTAATATAATCGAGGTGTTCCAGGATATAGATAGGGTACGTTGTGGAACCGTGCCGCGGGTATCAGCCGAGCGTGCTCTGTGCCAACGTGGTCTTCTGGCGCTTTTGTCATCGAGAGAAAGAGACACGCTCTACAAATGCTTCGGATACAGGCGAGGATGCGGTGACGAGGTAAATTATCGTGCACTGTGCAACGCTCTGGATGTACTGTACGCCACTACTAATGCTCAAAAATGTTAA
Protein
MVRDGIRPFDFLRQFDPRNELVIPRADFYRGLASAGLSLTPLEMDTLMEVFSAPGRRRYVEYARFSSTVGEALTQEGLERAPLLVPVPHLPTVDTPLNYLNFEERNIVSGALAKLAKYPDQLSNIIEVFQDIDRVRCGTVPRVSAERALCQRGLLALLSSRERDTLYKCFGYRRGCGDEVNYRALCNALDVLYATTNAQKC
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Similarity
Belongs to the MICOS complex subunit Mic60 family.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the G-protein coupled receptor 1 family.
Uniprot
H9JV83
A0A2A4JF51
A0A2W1BRS6
A0A212EHH3
A0A2H1X3K4
A0A3S2P1L2
+ More
A0A437ARA9 A0A194Q1X5 A0A0N1IH31 A0A0L7LNE7 A0A0T6B5X0 D6W947 N6U0C9 U4UF71 N6TXK3 A0A1W4WLJ6 A0A1Y1LDK1 A0A2P8YA30 A0A1S4FJD7 E0W159 A0A0L7RJB4 A0A1B6LR61 A0A182QSD8 A0A2A3EFG4 A0A0L0BYC0 A0A1B6MF68 B0WNC0 A0A087ZWP7 A0A026WHW5 A0A0J7KWR4 A0A1Q3FT57 A0A1B6K8A9 A0A154P491 A0A182TPE4 A0A2C9GR75 Q7QBY7 A0A182V3N9 A0A1B6DIZ4 A0A182VQA2 A0A1B6E3V0 A0A1I8PT42 A0A034VHG8 A0A084VJQ0 A0A1A9YS41 A0A1B0C3I7 A0A1B0FM66 A0A1B6E8U0 A0A1A9VKK1 A0A2R7VZZ3 A0A182XIC1 A0A195BFD8 A0A158NRF0 A0A182PA37 A0A3L8DJY8 A0A0K8W749 A0A0K8W8K1 A0A1A9ZG80 W8AHL7 A0A0K8V3S6 A0A182LFF7 A0A182S5M9 A0A1I8M395 A0A182MLL2 A0A0A1WR13 A0A151XF24 A0A182Y788 B4JIA6 A0A336KCV6 B4M0T2 A0A3B0KRI1 A0A336LCA6 A0A195FJH1 B4NKF0 A0A3B0KHD4 A0A1A9W6F7 E2AGH4 K7J532 A0A232ENR8 B4KA60 Q297Q1 B4G2V1 W5JW03 A0A182N4N9 B4PQG5 A0A182JYI6 B4R175 A0A182F6G5 B4HFQ9 B3P429 A0A1W4UR15 B3LXA1 A0A0M9A5F1 A0A0B4K6G7 Q9VFT7 A0A151JBN9 A0A182RCX1 A0A151IFQ2
A0A437ARA9 A0A194Q1X5 A0A0N1IH31 A0A0L7LNE7 A0A0T6B5X0 D6W947 N6U0C9 U4UF71 N6TXK3 A0A1W4WLJ6 A0A1Y1LDK1 A0A2P8YA30 A0A1S4FJD7 E0W159 A0A0L7RJB4 A0A1B6LR61 A0A182QSD8 A0A2A3EFG4 A0A0L0BYC0 A0A1B6MF68 B0WNC0 A0A087ZWP7 A0A026WHW5 A0A0J7KWR4 A0A1Q3FT57 A0A1B6K8A9 A0A154P491 A0A182TPE4 A0A2C9GR75 Q7QBY7 A0A182V3N9 A0A1B6DIZ4 A0A182VQA2 A0A1B6E3V0 A0A1I8PT42 A0A034VHG8 A0A084VJQ0 A0A1A9YS41 A0A1B0C3I7 A0A1B0FM66 A0A1B6E8U0 A0A1A9VKK1 A0A2R7VZZ3 A0A182XIC1 A0A195BFD8 A0A158NRF0 A0A182PA37 A0A3L8DJY8 A0A0K8W749 A0A0K8W8K1 A0A1A9ZG80 W8AHL7 A0A0K8V3S6 A0A182LFF7 A0A182S5M9 A0A1I8M395 A0A182MLL2 A0A0A1WR13 A0A151XF24 A0A182Y788 B4JIA6 A0A336KCV6 B4M0T2 A0A3B0KRI1 A0A336LCA6 A0A195FJH1 B4NKF0 A0A3B0KHD4 A0A1A9W6F7 E2AGH4 K7J532 A0A232ENR8 B4KA60 Q297Q1 B4G2V1 W5JW03 A0A182N4N9 B4PQG5 A0A182JYI6 B4R175 A0A182F6G5 B4HFQ9 B3P429 A0A1W4UR15 B3LXA1 A0A0M9A5F1 A0A0B4K6G7 Q9VFT7 A0A151JBN9 A0A182RCX1 A0A151IFQ2
Pubmed
19121390
28756777
22118469
26354079
26227816
18362917
+ More
19820115 23537049 28004739 29403074 20566863 26108605 24508170 12364791 14747013 17210077 25348373 24438588 21347285 30249741 24495485 20966253 25315136 25830018 25244985 17994087 20798317 20075255 28648823 15632085 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
19820115 23537049 28004739 29403074 20566863 26108605 24508170 12364791 14747013 17210077 25348373 24438588 21347285 30249741 24495485 20966253 25315136 25830018 25244985 17994087 20798317 20075255 28648823 15632085 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01037347
BABH01037348
NWSH01001644
PCG70591.1
KZ149979
PZC75857.1
+ More
AGBW02014893 OWR40921.1 ODYU01013136 SOQ59826.1 RSAL01000053 RVE50039.1 RSAL01003121 RVE40335.1 KQ459579 KPI99546.1 KQ459753 KPJ20165.1 JTDY01000546 KOB76721.1 LJIG01009616 KRT82724.1 KQ971312 EEZ98210.1 APGK01047442 APGK01047443 KB741081 ENN74041.1 KB632295 ERL91682.1 ENN74040.1 GEZM01058719 JAV71703.1 PYGN01000762 PSN41117.1 DS235866 EEB19365.1 KQ414580 KOC71062.1 GEBQ01013898 JAT26079.1 AXCN02000058 KZ288259 PBC30477.1 JRES01001160 KNC24966.1 GEBQ01005413 JAT34564.1 DS232010 EDS31574.1 KK107238 EZA54679.1 LBMM01002535 KMQ94694.1 GFDL01004317 JAV30728.1 GECU01000008 JAT07699.1 KQ434812 KZC06769.1 APCN01000432 AAAB01008859 EAA07680.5 GEDC01011627 JAS25671.1 GEDC01004696 JAS32602.1 GAKP01017939 JAC41013.1 ATLV01013846 KE524905 KFB38194.1 JXJN01024960 CCAG010009081 GEDC01002993 JAS34305.1 KK854205 PTY13064.1 KQ976504 KYM82924.1 ADTU01023858 ADTU01023859 QOIP01000007 RLU20681.1 GDHF01005358 JAI46956.1 GDHF01004848 JAI47466.1 GAMC01018597 GAMC01018595 JAB87960.1 GDHF01018798 JAI33516.1 AXCM01005074 GBXI01012798 JAD01494.1 KQ982205 KYQ58992.1 CH916369 EDV92987.1 UFQS01000220 UFQT01000220 SSX01558.1 SSX21938.1 CH940650 EDW67374.1 OUUW01000013 SPP87851.1 UFQS01003048 UFQT01003048 SSX15109.1 SSX34489.1 KQ981522 KYN40553.1 CH964272 EDW84080.1 SPP87850.1 GL439310 EFN67507.1 AAZX01000327 AAZX01000328 AAZX01014618 NNAY01003101 OXU19977.1 CH933806 EDW16735.1 CM000070 EAL28154.1 CH479179 EDW24146.1 ADMH02000291 ETN67104.1 CM000160 EDW97261.1 CM000364 EDX13052.1 CH480815 EDW42294.1 CH954181 EDV49203.1 CH902617 EDV41701.2 KQ435729 KOX77600.1 AE014297 AFH06405.1 AY058348 AAF54962.2 AAL13577.1 KQ979122 KYN22536.1 KQ977771 KYM99975.1
AGBW02014893 OWR40921.1 ODYU01013136 SOQ59826.1 RSAL01000053 RVE50039.1 RSAL01003121 RVE40335.1 KQ459579 KPI99546.1 KQ459753 KPJ20165.1 JTDY01000546 KOB76721.1 LJIG01009616 KRT82724.1 KQ971312 EEZ98210.1 APGK01047442 APGK01047443 KB741081 ENN74041.1 KB632295 ERL91682.1 ENN74040.1 GEZM01058719 JAV71703.1 PYGN01000762 PSN41117.1 DS235866 EEB19365.1 KQ414580 KOC71062.1 GEBQ01013898 JAT26079.1 AXCN02000058 KZ288259 PBC30477.1 JRES01001160 KNC24966.1 GEBQ01005413 JAT34564.1 DS232010 EDS31574.1 KK107238 EZA54679.1 LBMM01002535 KMQ94694.1 GFDL01004317 JAV30728.1 GECU01000008 JAT07699.1 KQ434812 KZC06769.1 APCN01000432 AAAB01008859 EAA07680.5 GEDC01011627 JAS25671.1 GEDC01004696 JAS32602.1 GAKP01017939 JAC41013.1 ATLV01013846 KE524905 KFB38194.1 JXJN01024960 CCAG010009081 GEDC01002993 JAS34305.1 KK854205 PTY13064.1 KQ976504 KYM82924.1 ADTU01023858 ADTU01023859 QOIP01000007 RLU20681.1 GDHF01005358 JAI46956.1 GDHF01004848 JAI47466.1 GAMC01018597 GAMC01018595 JAB87960.1 GDHF01018798 JAI33516.1 AXCM01005074 GBXI01012798 JAD01494.1 KQ982205 KYQ58992.1 CH916369 EDV92987.1 UFQS01000220 UFQT01000220 SSX01558.1 SSX21938.1 CH940650 EDW67374.1 OUUW01000013 SPP87851.1 UFQS01003048 UFQT01003048 SSX15109.1 SSX34489.1 KQ981522 KYN40553.1 CH964272 EDW84080.1 SPP87850.1 GL439310 EFN67507.1 AAZX01000327 AAZX01000328 AAZX01014618 NNAY01003101 OXU19977.1 CH933806 EDW16735.1 CM000070 EAL28154.1 CH479179 EDW24146.1 ADMH02000291 ETN67104.1 CM000160 EDW97261.1 CM000364 EDX13052.1 CH480815 EDW42294.1 CH954181 EDV49203.1 CH902617 EDV41701.2 KQ435729 KOX77600.1 AE014297 AFH06405.1 AY058348 AAF54962.2 AAL13577.1 KQ979122 KYN22536.1 KQ977771 KYM99975.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000037510 UP000007266 UP000019118 UP000030742 UP000192223 UP000245037 UP000009046 UP000053825 UP000075886 UP000242457 UP000037069 UP000002320 UP000005203 UP000053097 UP000036403 UP000076502 UP000075902 UP000075840 UP000007062 UP000075903 UP000075920 UP000095300 UP000030765 UP000092443 UP000092460 UP000092444 UP000078200 UP000076407 UP000078540 UP000005205 UP000075885 UP000279307 UP000092445 UP000075882 UP000075901 UP000095301 UP000075883 UP000075809 UP000076408 UP000001070 UP000008792 UP000268350 UP000078541 UP000007798 UP000091820 UP000000311 UP000002358 UP000215335 UP000009192 UP000001819 UP000008744 UP000000673 UP000075884 UP000002282 UP000075881 UP000000304 UP000069272 UP000001292 UP000008711 UP000192221 UP000007801 UP000053105 UP000000803 UP000078492 UP000075900 UP000078542
UP000037510 UP000007266 UP000019118 UP000030742 UP000192223 UP000245037 UP000009046 UP000053825 UP000075886 UP000242457 UP000037069 UP000002320 UP000005203 UP000053097 UP000036403 UP000076502 UP000075902 UP000075840 UP000007062 UP000075903 UP000075920 UP000095300 UP000030765 UP000092443 UP000092460 UP000092444 UP000078200 UP000076407 UP000078540 UP000005205 UP000075885 UP000279307 UP000092445 UP000075882 UP000075901 UP000095301 UP000075883 UP000075809 UP000076408 UP000001070 UP000008792 UP000268350 UP000078541 UP000007798 UP000091820 UP000000311 UP000002358 UP000215335 UP000009192 UP000001819 UP000008744 UP000000673 UP000075884 UP000002282 UP000075881 UP000000304 UP000069272 UP000001292 UP000008711 UP000192221 UP000007801 UP000053105 UP000000803 UP000078492 UP000075900 UP000078542
Interpro
IPR002048
EF_hand_dom
+ More
IPR011992 EF-hand-dom_pair
IPR036249 Thioredoxin-like_sf
IPR013766 Thioredoxin_domain
IPR036882 Alba-like_dom_sf
IPR002775 DNA/RNA-bd_Alba-like
IPR019133 Mt-IM_prot_Mitofilin
IPR018247 EF_Hand_1_Ca_BS
IPR001734 Na/solute_symporter
IPR038377 Na/Glc_symporter_sf
IPR013087 Znf_C2H2_type
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR011992 EF-hand-dom_pair
IPR036249 Thioredoxin-like_sf
IPR013766 Thioredoxin_domain
IPR036882 Alba-like_dom_sf
IPR002775 DNA/RNA-bd_Alba-like
IPR019133 Mt-IM_prot_Mitofilin
IPR018247 EF_Hand_1_Ca_BS
IPR001734 Na/solute_symporter
IPR038377 Na/Glc_symporter_sf
IPR013087 Znf_C2H2_type
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
Gene 3D
ProteinModelPortal
H9JV83
A0A2A4JF51
A0A2W1BRS6
A0A212EHH3
A0A2H1X3K4
A0A3S2P1L2
+ More
A0A437ARA9 A0A194Q1X5 A0A0N1IH31 A0A0L7LNE7 A0A0T6B5X0 D6W947 N6U0C9 U4UF71 N6TXK3 A0A1W4WLJ6 A0A1Y1LDK1 A0A2P8YA30 A0A1S4FJD7 E0W159 A0A0L7RJB4 A0A1B6LR61 A0A182QSD8 A0A2A3EFG4 A0A0L0BYC0 A0A1B6MF68 B0WNC0 A0A087ZWP7 A0A026WHW5 A0A0J7KWR4 A0A1Q3FT57 A0A1B6K8A9 A0A154P491 A0A182TPE4 A0A2C9GR75 Q7QBY7 A0A182V3N9 A0A1B6DIZ4 A0A182VQA2 A0A1B6E3V0 A0A1I8PT42 A0A034VHG8 A0A084VJQ0 A0A1A9YS41 A0A1B0C3I7 A0A1B0FM66 A0A1B6E8U0 A0A1A9VKK1 A0A2R7VZZ3 A0A182XIC1 A0A195BFD8 A0A158NRF0 A0A182PA37 A0A3L8DJY8 A0A0K8W749 A0A0K8W8K1 A0A1A9ZG80 W8AHL7 A0A0K8V3S6 A0A182LFF7 A0A182S5M9 A0A1I8M395 A0A182MLL2 A0A0A1WR13 A0A151XF24 A0A182Y788 B4JIA6 A0A336KCV6 B4M0T2 A0A3B0KRI1 A0A336LCA6 A0A195FJH1 B4NKF0 A0A3B0KHD4 A0A1A9W6F7 E2AGH4 K7J532 A0A232ENR8 B4KA60 Q297Q1 B4G2V1 W5JW03 A0A182N4N9 B4PQG5 A0A182JYI6 B4R175 A0A182F6G5 B4HFQ9 B3P429 A0A1W4UR15 B3LXA1 A0A0M9A5F1 A0A0B4K6G7 Q9VFT7 A0A151JBN9 A0A182RCX1 A0A151IFQ2
A0A437ARA9 A0A194Q1X5 A0A0N1IH31 A0A0L7LNE7 A0A0T6B5X0 D6W947 N6U0C9 U4UF71 N6TXK3 A0A1W4WLJ6 A0A1Y1LDK1 A0A2P8YA30 A0A1S4FJD7 E0W159 A0A0L7RJB4 A0A1B6LR61 A0A182QSD8 A0A2A3EFG4 A0A0L0BYC0 A0A1B6MF68 B0WNC0 A0A087ZWP7 A0A026WHW5 A0A0J7KWR4 A0A1Q3FT57 A0A1B6K8A9 A0A154P491 A0A182TPE4 A0A2C9GR75 Q7QBY7 A0A182V3N9 A0A1B6DIZ4 A0A182VQA2 A0A1B6E3V0 A0A1I8PT42 A0A034VHG8 A0A084VJQ0 A0A1A9YS41 A0A1B0C3I7 A0A1B0FM66 A0A1B6E8U0 A0A1A9VKK1 A0A2R7VZZ3 A0A182XIC1 A0A195BFD8 A0A158NRF0 A0A182PA37 A0A3L8DJY8 A0A0K8W749 A0A0K8W8K1 A0A1A9ZG80 W8AHL7 A0A0K8V3S6 A0A182LFF7 A0A182S5M9 A0A1I8M395 A0A182MLL2 A0A0A1WR13 A0A151XF24 A0A182Y788 B4JIA6 A0A336KCV6 B4M0T2 A0A3B0KRI1 A0A336LCA6 A0A195FJH1 B4NKF0 A0A3B0KHD4 A0A1A9W6F7 E2AGH4 K7J532 A0A232ENR8 B4KA60 Q297Q1 B4G2V1 W5JW03 A0A182N4N9 B4PQG5 A0A182JYI6 B4R175 A0A182F6G5 B4HFQ9 B3P429 A0A1W4UR15 B3LXA1 A0A0M9A5F1 A0A0B4K6G7 Q9VFT7 A0A151JBN9 A0A182RCX1 A0A151IFQ2
Ontologies
PANTHER
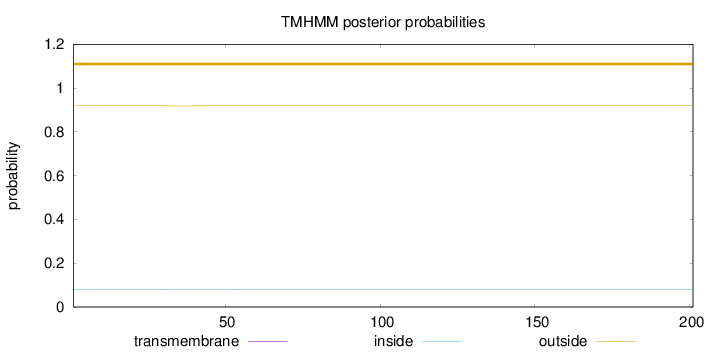
Topology
Subcellular location
Mitochondrion inner membrane
Length:
201
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01982
Exp number, first 60 AAs:
0.01892
Total prob of N-in:
0.08135
outside
1 - 201
Population Genetic Test Statistics
Pi
172.281094
Theta
170.418327
Tajima's D
0.019405
CLR
0.151791
CSRT
0.370031498425079
Interpretation
Uncertain
