Gene
KWMTBOMO03166
Pre Gene Modal
BGIBMGA013465
Annotation
high-affinity_choline_transporter_[Trichoplusia_ni]
Full name
High-affinity choline transporter 1
Location in the cell
PlasmaMembrane Reliability : 4.979
Sequence
CDS
ATGATAAACATAGCCGGCGTGATCTCCATCATTCTGTTTTACGTGCTGATCCTGGCAGTCGGTATATGGGCTGGGAGGAAGAAGCCTCCGGGAAATGACTCCGAAGAGGAGGTAATGCTGGCGGGACGATCCATTGGGCTGTTCGTCGGCATCTTTACTATGACTGCGACCTGGGTTGGTGGCGGCTATATCAATGGGACAGCAGAAGCGATATACACGTCGGGGCTGGTGTGGTGTCAGGCGCCTTTTGGATATGCCCTGTCCCTGGTCTTTGGCGGTATATTCTTCGCGAATCCAATGCGTCGTCAAGGCTACATTACGATGCTGGACCCTTTACAAGATTCTTTTGGAAGTCGCATGGGAGGTCTGCTGTTCTTGCCAGCTCTCTGCGGTGAGGTCTTTTGGGCTGCTGGAATATTGGCTGCGCTTGGTGCAACCCTGGCCGTCATCATCGACATGGATCAACGCACTTCTGTGATCTTCAGCGCATGCGTGGCAGTCTTCTACACTTTGTTCGGAGGCCTATACTCCGTGGCCTACACAGACGTCATTCAGTTATTCTGCATATTCATCGGTCTATGGATGTGTATACCGTTCGCTTGGGCGAACCCTCACGTGAAGCCGCTGAGCTCCATGGAGGTGGACTGGATAGGGAAGGTGGATTCGGACTATTATTGGTTCTATATCGATTATGGTCTATTGCTGATCTTTGGAGGGATTCCTTGGCAGGTTTATTTCCAACGAGTGTTGAGCAGCAAGACAGCAGGCAGAGCGCAGGTGCTCTCCTATGTAGCTGCAATTGGCTGCATCTTCATGGCCATCCCACCAGTTCTCATCGGGGCTATAGCTAAGGGCACTGCCTGGAACGAGACGGATTATCACGGAGACCTCACACCGGATGGAGAGCTCACCTCGGCGCAGACCTCAATGATTCTGCCACTGGTACTCCAACATCTCACACCGGAGTTTGTCTCATTCTTTGGGCTTGGAGCTGTCTCGGCTGCTGTCATGTCCTCGGCCGACTCGAGCGTGCTTAGTGCGTCTTCTATGTTTGCCAGGAATGTGTACAAGCTGATCTTCAGACAGAACGCTTCAGAGATGGAGATAATCTGGGTGATGCGGGTGTCTATACTTATCGTCGGGTTCCTGGCTACCGTCATGGCCCTCACCATACCTTCAATCTACGGATTATGGTCAATGTGCTCAGATCTGGTGTACGTGATTCTCTTCCCGCAACTCCTGATGGTGGTACACTTCAAGAAGTATTGCAACACCTATGGCTCTCTGGCTGCATACATCGTGGCTCTCTTAGTTAGGATGTCTGGAGGGGAGAAGATGTTAGGACTGCCGCCGCTGATCCACTATCCTATGTGGGACCCAAAAGAAGAAGTCCAGCTGTTTCCATTCCGAACCATGGCGATGGTTCTCTCACTCTTCACACTAGCCTTCATTTCATGGCTCTCCGTATTTCTCTTTAACTCCGGATACCTGAGCCCCGAGTCGGACTACTTCAACTGCGTCGTCAACATCCCGGAGGATATCCGTCGCGTGGACGAGCCATCCGAAGCTGGAGAGCAGATGTCCGTGCTGGGAGGAGTCCCGGGTCGACTCTACGGAGCTGCCTCCACTTTGGTCGGCCCCGATGAGAAGTCCGGAAGAATCAACCCGGCCCTGGAACCGGATGACGATGACCTGGATGACCCCAATGACGGGCCGACGCCGCTCTTTGGCTCCAGTAGCGCCCCGCCCCCAGTGCAACCCTACGGCAAGCAGACCGCCTTTTAA
Protein
MINIAGVISIILFYVLILAVGIWAGRKKPPGNDSEEEVMLAGRSIGLFVGIFTMTATWVGGGYINGTAEAIYTSGLVWCQAPFGYALSLVFGGIFFANPMRRQGYITMLDPLQDSFGSRMGGLLFLPALCGEVFWAAGILAALGATLAVIIDMDQRTSVIFSACVAVFYTLFGGLYSVAYTDVIQLFCIFIGLWMCIPFAWANPHVKPLSSMEVDWIGKVDSDYYWFYIDYGLLLIFGGIPWQVYFQRVLSSKTAGRAQVLSYVAAIGCIFMAIPPVLIGAIAKGTAWNETDYHGDLTPDGELTSAQTSMILPLVLQHLTPEFVSFFGLGAVSAAVMSSADSSVLSASSMFARNVYKLIFRQNASEMEIIWVMRVSILIVGFLATVMALTIPSIYGLWSMCSDLVYVILFPQLLMVVHFKKYCNTYGSLAAYIVALLVRMSGGEKMLGLPPLIHYPMWDPKEEVQLFPFRTMAMVLSLFTLAFISWLSVFLFNSGYLSPESDYFNCVVNIPEDIRRVDEPSEAGEQMSVLGGVPGRLYGAASTLVGPDEKSGRINPALEPDDDDLDDPNDGPTPLFGSSSAPPPVQPYGKQTAF
Summary
Description
Imports choline from the extracellular space to the neuron with high affinity. Rate-limiting step in acetylcholine synthesis. Sodium ion and chloride ion dependent (By similarity).
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Keywords
Complete proteome
Glycoprotein
Ion transport
Membrane
Neurotransmitter biosynthesis
Reference proteome
Sodium
Sodium transport
Symport
Transmembrane
Transmembrane helix
Transport
Feature
chain High-affinity choline transporter 1
Uniprot
A0A212FCP7
A0A2A4JEV3
A0A2W1BT95
Q693W0
A0A194Q2I4
A0A0N0PEZ3
+ More
A0A0G3VIB0 A0A0L7LMF2 W8C9U8 A0A0K8UFU7 B4NI91 B4KC51 A0A182UTY4 A0A182IUT4 A0A182Y786 A0A182LFE7 A0A182XIB9 Q7QBY8 A0A182HTA4 A0A182PA39 A0A182W2X7 A0A182Q9L8 A0A0M4EGX8 A0A1A9XDK5 A0A1B0BTV7 A0A0L0CQ40 A0A1A9VS46 B4JS82 A0A1A9ZGD7 Q299V8 A0A336MDH6 B4MBB9 B3P3C1 B4QTZ8 B4PM04 Q9VE46 A0A1W4UB27 A0A182LS72 B3M1V6 A0A1I8MHF5 A0A1J1IZV3 A0A1B0CEQ2 A0A3B0JDD0 A0A1I8PBJ6 A0A182N4P0 A0A182GZL3 E0W160 A0A195BEB5 A0A151IFN6 F4WAY2 A0A158NRC3 A0A2J7Q7E1 A0A151XF84 B0W360 A0A151JBP3 A0A1B6KPI3 A0A1B6KJK3 A0A195FJW5 E9IS26 A0A3L8DJT7 A0A026WFH4 A0A084W1K7 A0A0L7RJP5 K7J532 A0A067R0S6 V9ICA4 A0A2A3EFI4 A0A1B6E374 A0A087ZWP8 A0A310STY2 B4I262 A0A0M9A7S1 A0A0C9QJM2 A0A154P4I0 A0A232EQX6 A0A0A9ZFC7 A0A1W4WRS7 A0A182F6G4 D6W957 W5JSJ8 E2AGH2 A0A1B6EBI2 J9KAD4 A0A2H8THZ3 A0A2S2Q3S9 E9G3N6 A0A0P5DK19 A0A0P6FYM0 A0A336JYC9 A0A0P6IIM9 A0A182TVE5 A0A182RYG9 A0A1D2N665 A0A3S3PK89 Q16YT6 A0A132A6Y8 A0A1V9XZS3 A0A147BPX9
A0A0G3VIB0 A0A0L7LMF2 W8C9U8 A0A0K8UFU7 B4NI91 B4KC51 A0A182UTY4 A0A182IUT4 A0A182Y786 A0A182LFE7 A0A182XIB9 Q7QBY8 A0A182HTA4 A0A182PA39 A0A182W2X7 A0A182Q9L8 A0A0M4EGX8 A0A1A9XDK5 A0A1B0BTV7 A0A0L0CQ40 A0A1A9VS46 B4JS82 A0A1A9ZGD7 Q299V8 A0A336MDH6 B4MBB9 B3P3C1 B4QTZ8 B4PM04 Q9VE46 A0A1W4UB27 A0A182LS72 B3M1V6 A0A1I8MHF5 A0A1J1IZV3 A0A1B0CEQ2 A0A3B0JDD0 A0A1I8PBJ6 A0A182N4P0 A0A182GZL3 E0W160 A0A195BEB5 A0A151IFN6 F4WAY2 A0A158NRC3 A0A2J7Q7E1 A0A151XF84 B0W360 A0A151JBP3 A0A1B6KPI3 A0A1B6KJK3 A0A195FJW5 E9IS26 A0A3L8DJT7 A0A026WFH4 A0A084W1K7 A0A0L7RJP5 K7J532 A0A067R0S6 V9ICA4 A0A2A3EFI4 A0A1B6E374 A0A087ZWP8 A0A310STY2 B4I262 A0A0M9A7S1 A0A0C9QJM2 A0A154P4I0 A0A232EQX6 A0A0A9ZFC7 A0A1W4WRS7 A0A182F6G4 D6W957 W5JSJ8 E2AGH2 A0A1B6EBI2 J9KAD4 A0A2H8THZ3 A0A2S2Q3S9 E9G3N6 A0A0P5DK19 A0A0P6FYM0 A0A336JYC9 A0A0P6IIM9 A0A182TVE5 A0A182RYG9 A0A1D2N665 A0A3S3PK89 Q16YT6 A0A132A6Y8 A0A1V9XZS3 A0A147BPX9
Pubmed
22118469
28756777
15607656
26354079
26227816
24495485
+ More
17994087 25244985 20966253 12364791 14747013 17210077 26108605 15632085 18057021 17550304 10731132 12537572 12537569 25315136 26483478 20566863 21719571 21347285 21282665 30249741 24508170 24438588 20075255 24845553 28648823 25401762 18362917 19820115 20920257 23761445 20798317 21292972 27289101 17510324 26555130 28327890 29652888
17994087 25244985 20966253 12364791 14747013 17210077 26108605 15632085 18057021 17550304 10731132 12537572 12537569 25315136 26483478 20566863 21719571 21347285 21282665 30249741 24508170 24438588 20075255 24845553 28648823 25401762 18362917 19820115 20920257 23761445 20798317 21292972 27289101 17510324 26555130 28327890 29652888
EMBL
AGBW02009155
OWR51514.1
NWSH01001644
PCG70587.1
KZ149979
PZC75856.1
+ More
AY629593 AAT88074.1 KQ459579 KPI99548.1 KQ459753 KPJ20167.1 KP657648 AKL78873.1 JTDY01000546 KOB76718.1 GAMC01006336 JAC00220.1 GDHF01026886 JAI25428.1 CH964272 EDW83673.1 CH933806 EDW16924.1 AAAB01008859 EAA07459.3 APCN01000429 APCN01000430 APCN01000431 AXCN02000058 AXCN02000059 CP012526 ALC45471.1 JXJN01020370 JXJN01020371 JRES01000062 KNC34473.1 CH916373 EDV94622.1 CM000070 EAL27593.1 UFQT01001009 SSX28452.1 CH940656 EDW58390.1 KRF78256.1 CH954181 EDV48573.1 CM000364 EDX12421.1 CM000160 EDW95936.1 AE014297 AY047521 AXCM01000096 CH902617 EDV42216.1 CVRI01000065 CRL05795.1 AJWK01009066 AJWK01009067 OUUW01000005 SPP80384.1 JXUM01099816 JXUM01099817 KQ564521 KXJ72141.1 DS235866 EEB19366.1 KQ976504 KYM82926.1 KQ977771 KYM99974.1 GL888056 EGI68602.1 ADTU01023859 ADTU01023860 NEVH01017443 PNF24499.1 KQ982205 KYQ58991.1 DS231831 EDS31203.1 KQ979122 KYN22537.1 GEBQ01026853 JAT13124.1 GEBQ01028372 JAT11605.1 KQ981522 KYN40552.1 GL765283 EFZ16638.1 QOIP01000007 RLU20680.1 KK107238 EZA54678.1 ATLV01019372 ATLV01019373 KE525269 KFB44101.1 KQ414580 KOC71059.1 AAZX01000327 AAZX01000328 AAZX01014618 KK852798 KDR16364.1 JR039569 AEY58758.1 KZ288259 PBC30478.1 GEDC01004915 JAS32383.1 KQ760497 OAD60101.1 CH480820 EDW54619.1 KQ435729 KOX77599.1 GBYB01000757 JAG70524.1 KQ434812 KZC06767.1 NNAY01002694 OXU20760.1 GBHO01000380 JAG43224.1 KQ971312 EEZ98445.2 ADMH02000291 ETN67106.1 GL439310 EFN67505.1 GEDC01002025 JAS35273.1 ABLF02030995 GFXV01001921 MBW13726.1 GGMS01002659 MBY71862.1 GL732531 EFX85951.1 GDIP01156267 LRGB01001574 JAJ67135.1 KZS11610.1 GDIQ01041106 JAN53631.1 UFQS01000013 UFQT01000013 SSW97241.1 SSX17627.1 GDIQ01021329 JAN73408.1 LJIJ01000187 ODN00758.1 NCKU01000774 NCKU01000257 RWS14213.1 RWS16282.1 CH477509 EAT39796.1 JXLN01011066 KPM06722.1 MNPL01001609 OQR78985.1 GEGO01002595 JAR92809.1
AY629593 AAT88074.1 KQ459579 KPI99548.1 KQ459753 KPJ20167.1 KP657648 AKL78873.1 JTDY01000546 KOB76718.1 GAMC01006336 JAC00220.1 GDHF01026886 JAI25428.1 CH964272 EDW83673.1 CH933806 EDW16924.1 AAAB01008859 EAA07459.3 APCN01000429 APCN01000430 APCN01000431 AXCN02000058 AXCN02000059 CP012526 ALC45471.1 JXJN01020370 JXJN01020371 JRES01000062 KNC34473.1 CH916373 EDV94622.1 CM000070 EAL27593.1 UFQT01001009 SSX28452.1 CH940656 EDW58390.1 KRF78256.1 CH954181 EDV48573.1 CM000364 EDX12421.1 CM000160 EDW95936.1 AE014297 AY047521 AXCM01000096 CH902617 EDV42216.1 CVRI01000065 CRL05795.1 AJWK01009066 AJWK01009067 OUUW01000005 SPP80384.1 JXUM01099816 JXUM01099817 KQ564521 KXJ72141.1 DS235866 EEB19366.1 KQ976504 KYM82926.1 KQ977771 KYM99974.1 GL888056 EGI68602.1 ADTU01023859 ADTU01023860 NEVH01017443 PNF24499.1 KQ982205 KYQ58991.1 DS231831 EDS31203.1 KQ979122 KYN22537.1 GEBQ01026853 JAT13124.1 GEBQ01028372 JAT11605.1 KQ981522 KYN40552.1 GL765283 EFZ16638.1 QOIP01000007 RLU20680.1 KK107238 EZA54678.1 ATLV01019372 ATLV01019373 KE525269 KFB44101.1 KQ414580 KOC71059.1 AAZX01000327 AAZX01000328 AAZX01014618 KK852798 KDR16364.1 JR039569 AEY58758.1 KZ288259 PBC30478.1 GEDC01004915 JAS32383.1 KQ760497 OAD60101.1 CH480820 EDW54619.1 KQ435729 KOX77599.1 GBYB01000757 JAG70524.1 KQ434812 KZC06767.1 NNAY01002694 OXU20760.1 GBHO01000380 JAG43224.1 KQ971312 EEZ98445.2 ADMH02000291 ETN67106.1 GL439310 EFN67505.1 GEDC01002025 JAS35273.1 ABLF02030995 GFXV01001921 MBW13726.1 GGMS01002659 MBY71862.1 GL732531 EFX85951.1 GDIP01156267 LRGB01001574 JAJ67135.1 KZS11610.1 GDIQ01041106 JAN53631.1 UFQS01000013 UFQT01000013 SSW97241.1 SSX17627.1 GDIQ01021329 JAN73408.1 LJIJ01000187 ODN00758.1 NCKU01000774 NCKU01000257 RWS14213.1 RWS16282.1 CH477509 EAT39796.1 JXLN01011066 KPM06722.1 MNPL01001609 OQR78985.1 GEGO01002595 JAR92809.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
UP000007798
+ More
UP000009192 UP000075903 UP000075880 UP000076408 UP000075882 UP000076407 UP000007062 UP000075840 UP000075885 UP000075920 UP000075886 UP000092553 UP000092443 UP000092460 UP000037069 UP000078200 UP000001070 UP000092445 UP000001819 UP000008792 UP000008711 UP000000304 UP000002282 UP000000803 UP000192221 UP000075883 UP000007801 UP000095301 UP000183832 UP000092461 UP000268350 UP000095300 UP000075884 UP000069940 UP000249989 UP000009046 UP000078540 UP000078542 UP000007755 UP000005205 UP000235965 UP000075809 UP000002320 UP000078492 UP000078541 UP000279307 UP000053097 UP000030765 UP000053825 UP000002358 UP000027135 UP000242457 UP000005203 UP000001292 UP000053105 UP000076502 UP000215335 UP000192223 UP000069272 UP000007266 UP000000673 UP000000311 UP000007819 UP000000305 UP000076858 UP000075902 UP000075900 UP000094527 UP000285301 UP000008820 UP000192247
UP000009192 UP000075903 UP000075880 UP000076408 UP000075882 UP000076407 UP000007062 UP000075840 UP000075885 UP000075920 UP000075886 UP000092553 UP000092443 UP000092460 UP000037069 UP000078200 UP000001070 UP000092445 UP000001819 UP000008792 UP000008711 UP000000304 UP000002282 UP000000803 UP000192221 UP000075883 UP000007801 UP000095301 UP000183832 UP000092461 UP000268350 UP000095300 UP000075884 UP000069940 UP000249989 UP000009046 UP000078540 UP000078542 UP000007755 UP000005205 UP000235965 UP000075809 UP000002320 UP000078492 UP000078541 UP000279307 UP000053097 UP000030765 UP000053825 UP000002358 UP000027135 UP000242457 UP000005203 UP000001292 UP000053105 UP000076502 UP000215335 UP000192223 UP000069272 UP000007266 UP000000673 UP000000311 UP000007819 UP000000305 UP000076858 UP000075902 UP000075900 UP000094527 UP000285301 UP000008820 UP000192247
PRIDE
Pfam
PF00474 SSF
Interpro
SUPFAM
SSF47473
SSF47473
Gene 3D
ProteinModelPortal
A0A212FCP7
A0A2A4JEV3
A0A2W1BT95
Q693W0
A0A194Q2I4
A0A0N0PEZ3
+ More
A0A0G3VIB0 A0A0L7LMF2 W8C9U8 A0A0K8UFU7 B4NI91 B4KC51 A0A182UTY4 A0A182IUT4 A0A182Y786 A0A182LFE7 A0A182XIB9 Q7QBY8 A0A182HTA4 A0A182PA39 A0A182W2X7 A0A182Q9L8 A0A0M4EGX8 A0A1A9XDK5 A0A1B0BTV7 A0A0L0CQ40 A0A1A9VS46 B4JS82 A0A1A9ZGD7 Q299V8 A0A336MDH6 B4MBB9 B3P3C1 B4QTZ8 B4PM04 Q9VE46 A0A1W4UB27 A0A182LS72 B3M1V6 A0A1I8MHF5 A0A1J1IZV3 A0A1B0CEQ2 A0A3B0JDD0 A0A1I8PBJ6 A0A182N4P0 A0A182GZL3 E0W160 A0A195BEB5 A0A151IFN6 F4WAY2 A0A158NRC3 A0A2J7Q7E1 A0A151XF84 B0W360 A0A151JBP3 A0A1B6KPI3 A0A1B6KJK3 A0A195FJW5 E9IS26 A0A3L8DJT7 A0A026WFH4 A0A084W1K7 A0A0L7RJP5 K7J532 A0A067R0S6 V9ICA4 A0A2A3EFI4 A0A1B6E374 A0A087ZWP8 A0A310STY2 B4I262 A0A0M9A7S1 A0A0C9QJM2 A0A154P4I0 A0A232EQX6 A0A0A9ZFC7 A0A1W4WRS7 A0A182F6G4 D6W957 W5JSJ8 E2AGH2 A0A1B6EBI2 J9KAD4 A0A2H8THZ3 A0A2S2Q3S9 E9G3N6 A0A0P5DK19 A0A0P6FYM0 A0A336JYC9 A0A0P6IIM9 A0A182TVE5 A0A182RYG9 A0A1D2N665 A0A3S3PK89 Q16YT6 A0A132A6Y8 A0A1V9XZS3 A0A147BPX9
A0A0G3VIB0 A0A0L7LMF2 W8C9U8 A0A0K8UFU7 B4NI91 B4KC51 A0A182UTY4 A0A182IUT4 A0A182Y786 A0A182LFE7 A0A182XIB9 Q7QBY8 A0A182HTA4 A0A182PA39 A0A182W2X7 A0A182Q9L8 A0A0M4EGX8 A0A1A9XDK5 A0A1B0BTV7 A0A0L0CQ40 A0A1A9VS46 B4JS82 A0A1A9ZGD7 Q299V8 A0A336MDH6 B4MBB9 B3P3C1 B4QTZ8 B4PM04 Q9VE46 A0A1W4UB27 A0A182LS72 B3M1V6 A0A1I8MHF5 A0A1J1IZV3 A0A1B0CEQ2 A0A3B0JDD0 A0A1I8PBJ6 A0A182N4P0 A0A182GZL3 E0W160 A0A195BEB5 A0A151IFN6 F4WAY2 A0A158NRC3 A0A2J7Q7E1 A0A151XF84 B0W360 A0A151JBP3 A0A1B6KPI3 A0A1B6KJK3 A0A195FJW5 E9IS26 A0A3L8DJT7 A0A026WFH4 A0A084W1K7 A0A0L7RJP5 K7J532 A0A067R0S6 V9ICA4 A0A2A3EFI4 A0A1B6E374 A0A087ZWP8 A0A310STY2 B4I262 A0A0M9A7S1 A0A0C9QJM2 A0A154P4I0 A0A232EQX6 A0A0A9ZFC7 A0A1W4WRS7 A0A182F6G4 D6W957 W5JSJ8 E2AGH2 A0A1B6EBI2 J9KAD4 A0A2H8THZ3 A0A2S2Q3S9 E9G3N6 A0A0P5DK19 A0A0P6FYM0 A0A336JYC9 A0A0P6IIM9 A0A182TVE5 A0A182RYG9 A0A1D2N665 A0A3S3PK89 Q16YT6 A0A132A6Y8 A0A1V9XZS3 A0A147BPX9
PDB
2XQ2
E-value=2.76757e-07,
Score=133
Ontologies
GO
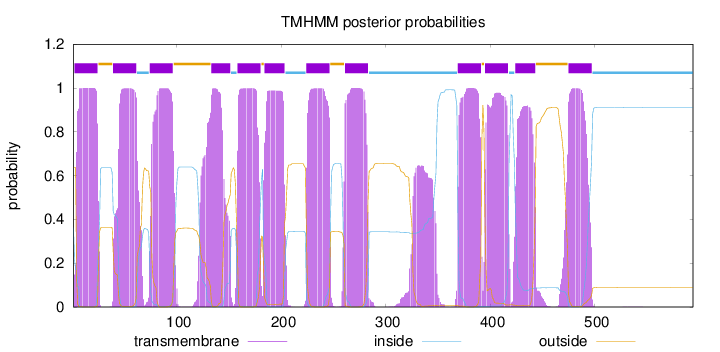
Topology
Subcellular location
Membrane
Length:
594
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
275.68105
Exp number, first 60 AAs:
40.04532
Total prob of N-in:
0.36286
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 24
outside
25 - 38
TMhelix
39 - 61
inside
62 - 73
TMhelix
74 - 96
outside
97 - 132
TMhelix
133 - 151
inside
152 - 157
TMhelix
158 - 180
outside
181 - 183
TMhelix
184 - 203
inside
204 - 223
TMhelix
224 - 246
outside
247 - 260
TMhelix
261 - 283
inside
284 - 368
TMhelix
369 - 391
outside
392 - 394
TMhelix
395 - 417
inside
418 - 423
TMhelix
424 - 443
outside
444 - 474
TMhelix
475 - 497
inside
498 - 594
Population Genetic Test Statistics
Pi
176.829002
Theta
155.096015
Tajima's D
0.564219
CLR
0.317967
CSRT
0.535523223838808
Interpretation
Uncertain
