Gene
KWMTBOMO03164
Pre Gene Modal
BGIBMGA013466
Annotation
Vesicular_acetylcholine_transporter_[Papilio_xuthus]
Full name
Vesicular acetylcholine transporter
Location in the cell
PlasmaMembrane Reliability : 4.643
Sequence
CDS
ATGGCGGAGGGACCACAGACGATATGGCAGAGGATCGACAATTCCATGATTCCAATCGTCAACTTGGAAGTGAGGGAAGTAAGGGAGATATTATGGGAGAAAATCAAAGAGCCGACCTCCCAGAGAAAAATCATCCTCGTTATAGTATCTATAGCCTTGTTGCTAGACAACATGTTGTATATGGTCATAGTGCCAATAATCCCAGACTATCTGAGATACATTGGCGCATGGGGTGAGGCAGCTTACGACCACGTGGTTACTTTGCCGCCAATCAAAGAAGGGAACAGAACAGTGATACCGACGAAGATCATTCCTGGTTCTCATGAGGGGCAAGATTCGGCTACTGGAGTTCTGTTTGCATCAAAAGCTATAGTTCAGCTGATGATTAATCCCTTTTCCGGTGCTTTAATCGATCGTATCGGCTACGATATTCCCATGATGATAGGACTGATCATTATGTTCCTGTCGACATCGATCTTCGCTTGCGGTAGAAGTTATAGCATGTTGTTCTTTGCTAGGAGTCTCCAAGGGGTCGGATCGGCGTTTGCTGATACATCAGGCTTGGCTATGATCGCTGACAGATTTACCGAAGAATCCGAGCGCTCTAAAGCACTAGGCATAGCACTGGCATTTATTAGTTTTGGAAGCCTGGTCGCTCCACCATTTGGCGGTGCTTTATATCAATTTGCGGGCAAGGAAGTGCCATTTCTTATTTTGGCATTGATATCCTTAATGGATGGATTCATGTTATTACTCGTCATGAAGCCAATAAAGACTCAGATCAAAGAGGCGAATGAACCAAAACCAGCTGGAACTCCAATTTGGAAGCTCTTGATTGATCCTTACATCGCAGTTTGCGCTGGAGCTTTAATGATGTCGAATGTAGCGCTTGCATTCCTAGAGCCCACGATTTCATCTTGGATGGAAGACAATTTGACTAAAGATAATTGGAAAATAGGCATGATATGGCTACCGGCTTTCTTTCCTCATGTTCTGGGTGTGATCATAACCGTCAAGATGGCAAAGAAGTACCCTCAACACCAATGGTTGATGGCTGCTGGTGGGTTGGCTCTGGAAGGTTTATGTTGTTTCATCATACCTTTCGCGAGTTCGTATAAGATGTTGATGATTCCTATTTGCGGTATATGCTTCGGGATTGCCTTGATCGACACAGCTTTGCTGCCAACTCTTGGATACTTGGTTGATGTTCGTTATGTATCTGTCTACGGAAGTATTTATGCCATTGCTGACATATCATACTCCTTCGCTTATGCTGTAGGACCCATCATCGCTGGAGAAGTCGTCGAGGCTATTGGATTCACTGCTTTGAACCTGCTAATTGCGTTCAGTAATCTTCTTTATGCACCAGTGCTGATGTACCTTCGGCATATTTATGACTTCAAGCCATTTGAGAACGAAGCCAATATTTTGATGGCTGATCCACCAGATAAAGAATACCAGACTTACACCATGCAAGACCAGAAGCCGATTAATGGAGACTATAAAAATCACTTGGAGTACAATAGTAGTGCCGGTCAAGCTACTCAGGAATCAAATGTGGATGCTGGGAATTATTCTTACGACCAGACGTATCAAGGAGGAAGTTATAGCCAAGGCTATGAACAGCAAGGATACCATCAAGAATATCAGCAGGCGGAGTACAACCAACCGAGGCAGTTACCGGCTCAGCCACAGACGGATTATGAATACGAAGCATAG
Protein
MAEGPQTIWQRIDNSMIPIVNLEVREVREILWEKIKEPTSQRKIILVIVSIALLLDNMLYMVIVPIIPDYLRYIGAWGEAAYDHVVTLPPIKEGNRTVIPTKIIPGSHEGQDSATGVLFASKAIVQLMINPFSGALIDRIGYDIPMMIGLIIMFLSTSIFACGRSYSMLFFARSLQGVGSAFADTSGLAMIADRFTEESERSKALGIALAFISFGSLVAPPFGGALYQFAGKEVPFLILALISLMDGFMLLLVMKPIKTQIKEANEPKPAGTPIWKLLIDPYIAVCAGALMMSNVALAFLEPTISSWMEDNLTKDNWKIGMIWLPAFFPHVLGVIITVKMAKKYPQHQWLMAAGGLALEGLCCFIIPFASSYKMLMIPICGICFGIALIDTALLPTLGYLVDVRYVSVYGSIYAIADISYSFAYAVGPIIAGEVVEAIGFTALNLLIAFSNLLYAPVLMYLRHIYDFKPFENEANILMADPPDKEYQTYTMQDQKPINGDYKNHLEYNSSAGQATQESNVDAGNYSYDQTYQGGSYSQGYEQQGYHQEYQQAEYNQPRQLPAQPQTDYEYEA
Summary
Similarity
Belongs to the major facilitator superfamily. Vesicular transporter family.
Keywords
Complete proteome
Glycoprotein
Membrane
Neurotransmitter transport
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Vesicular acetylcholine transporter
Uniprot
H9JVA3
A0A212FCP6
A0A0N0PEL3
A0A194Q1Y0
A0A2H1VZT8
A0A2A4J2I7
+ More
A0A0G3VG09 A0A0L7LF40 D6W971 A0A2J7Q7C1 A0A2P8YA25 A0A182Y784 A0A067RCA1 A0A1Y1KTX3 A0A182RYG7 A0A084W1L3 A0A182J1B3 A0A182N4P3 A0A182M1V9 T1I654 A0A1J1J2W2 A0A182W2X5 A0A0T6BDE1 U5EX12 A0A182PA41 A0A182UE01 A0A2R7WRG2 A0A182UGY9 Q16SX3 Q7PR83 A0A182XIB7 A0A182T7R7 E0W161 A0A182G771 A0A0L0CQ39 A0A182VCH4 E2AGG8 A0A182F6G2 W5JVM7 F4WB43 A0A1S3D026 A0A232EMV3 K7J534 A0A195FJG4 A0A1A9XZF5 A0A1B0BUL9 A0A1A9VIY3 E2C7G8 A0A1A9ZL63 A0A1B0GET2 A0A310SPT4 A0A1A9W5V3 E9IS27 A0A336MP64 A0A0L7RJK5 A0A195BFI5 A0A087ZWQ0 A0A158NRC4 A0A1W4X2P3 A0A1I8NUP2 A0A151JBS7 A0A2A3EGY5 A0A1I8M3G4 A0A026WF51 V9I6X8 A0A182KBJ2 B0W363 A0A0J7L6J2 A0A0C9RXM3 A0A182QP81 W8C1Q4 A0A0Q9WUM0 A0A154P5Y7 A0A1W4UPF5 A0A3B0K361 A0A182HTA2 A0A0Q9X4V4 A0A0M5J8U6 B4K1S6 B4MBC3 I5ANM2 A0A0P8XY20 A0A2H8TD87 D2NUG3 O17444 A0A2S2NLL7 J9JXZ7 A0A2S2QJZ0 E9G3N1 B7QD10 A0A0P5XI13 A0A0P6G6T8 A0A1D2N677 A0A0P6C1V8 A0A2I6QBP8 A0A226CYU2 A0A0P5B8W8 A0A0P5BAW4 A0A0P5IXP8 A0A0P5KY64
A0A0G3VG09 A0A0L7LF40 D6W971 A0A2J7Q7C1 A0A2P8YA25 A0A182Y784 A0A067RCA1 A0A1Y1KTX3 A0A182RYG7 A0A084W1L3 A0A182J1B3 A0A182N4P3 A0A182M1V9 T1I654 A0A1J1J2W2 A0A182W2X5 A0A0T6BDE1 U5EX12 A0A182PA41 A0A182UE01 A0A2R7WRG2 A0A182UGY9 Q16SX3 Q7PR83 A0A182XIB7 A0A182T7R7 E0W161 A0A182G771 A0A0L0CQ39 A0A182VCH4 E2AGG8 A0A182F6G2 W5JVM7 F4WB43 A0A1S3D026 A0A232EMV3 K7J534 A0A195FJG4 A0A1A9XZF5 A0A1B0BUL9 A0A1A9VIY3 E2C7G8 A0A1A9ZL63 A0A1B0GET2 A0A310SPT4 A0A1A9W5V3 E9IS27 A0A336MP64 A0A0L7RJK5 A0A195BFI5 A0A087ZWQ0 A0A158NRC4 A0A1W4X2P3 A0A1I8NUP2 A0A151JBS7 A0A2A3EGY5 A0A1I8M3G4 A0A026WF51 V9I6X8 A0A182KBJ2 B0W363 A0A0J7L6J2 A0A0C9RXM3 A0A182QP81 W8C1Q4 A0A0Q9WUM0 A0A154P5Y7 A0A1W4UPF5 A0A3B0K361 A0A182HTA2 A0A0Q9X4V4 A0A0M5J8U6 B4K1S6 B4MBC3 I5ANM2 A0A0P8XY20 A0A2H8TD87 D2NUG3 O17444 A0A2S2NLL7 J9JXZ7 A0A2S2QJZ0 E9G3N1 B7QD10 A0A0P5XI13 A0A0P6G6T8 A0A1D2N677 A0A0P6C1V8 A0A2I6QBP8 A0A226CYU2 A0A0P5B8W8 A0A0P5BAW4 A0A0P5IXP8 A0A0P5KY64
Pubmed
19121390
22118469
26354079
26227816
18362917
19820115
+ More
29403074 25244985 24845553 28004739 24438588 17510324 12364791 14747013 17210077 20566863 26483478 26108605 20798317 20920257 23761445 21719571 28648823 20075255 21282665 21347285 25315136 24508170 30249741 24495485 17994087 15632085 9446576 10731132 12537572 21292972 27289101 29236686
29403074 25244985 24845553 28004739 24438588 17510324 12364791 14747013 17210077 20566863 26483478 26108605 20798317 20920257 23761445 21719571 28648823 20075255 21282665 21347285 25315136 24508170 30249741 24495485 17994087 15632085 9446576 10731132 12537572 21292972 27289101 29236686
EMBL
BABH01037362
AB871890
BAO23492.1
AGBW02009155
OWR51511.1
KQ459753
+ More
KPJ20169.1 KQ459579 KPI99551.1 ODYU01005161 SOQ45754.1 NWSH01003475 PCG66221.1 KP657649 AKL78874.1 JTDY01001459 KOB73816.1 KQ971312 EEZ98455.2 NEVH01017443 PNF24483.1 PYGN01000762 PSN41105.1 KK852798 KDR16361.1 GEZM01075679 JAV64018.1 ATLV01019381 KE525269 KFB44107.1 AXCM01000067 ACPB03008308 CVRI01000065 CRL05790.1 LJIG01001596 KRT85344.1 GANO01001162 JAB58709.1 KK855354 PTY22144.1 CH477664 EAT37546.1 AAAB01008859 EAA07682.6 DS235866 EEB19367.1 JXUM01046136 KQ561464 KXJ78489.1 JRES01000062 KNC34475.1 GL439310 EFN67501.1 ADMH02000344 ETN66889.1 GL888056 EGI68598.1 NNAY01003307 OXU19647.1 AAZX01000328 KQ981522 KYN40548.1 JXJN01020792 GL453369 EFN76155.1 CCAG010012585 KQ760497 OAD60097.1 GL765283 EFZ16652.1 UFQS01001326 UFQT01001326 SSX10359.1 SSX30047.1 KQ414580 KOC71055.1 KQ976504 KYM82930.1 ADTU01023865 KQ979122 KYN22540.1 KZ288259 PBC30479.1 KK107238 QOIP01000007 EZA54672.1 RLU20763.1 JR035965 JR035966 JR035967 JR035968 AEY56830.1 AEY56831.1 AEY56832.1 AEY56833.1 DS231831 EDS31206.1 LBMM01000504 KMQ98193.1 GBYB01013930 JAG83697.1 AXCN02000061 GAMC01006914 JAB99641.1 CH964272 KRF99855.1 KQ434812 KZC06764.1 OUUW01000005 SPP80389.1 APCN01000427 CH933806 KRG02377.1 CP012526 ALC45474.1 CH917801 EDW04624.1 CH940656 EDW58394.1 CM000070 EIM52557.1 CH902617 KPU79602.1 GFXV01000249 MBW12054.1 BT120106 ADB13439.1 AF030197 AE014297 GGMR01005464 MBY18083.1 ABLF02025153 ABLF02025155 GGMS01008647 MBY77850.1 GL732531 EFX85953.1 ABJB011139869 DS910653 EEC16732.1 GDIP01071795 LRGB01001574 JAM31920.1 KZS11615.1 GDIQ01048214 JAN46523.1 LJIJ01000187 ODN00757.1 GDIP01010637 JAM93078.1 KY809750 AUN27676.1 LNIX01000063 OXA37206.1 GDIP01187824 JAJ35578.1 GDIP01187823 JAJ35579.1 GDIQ01207956 JAK43769.1 GDIQ01203454 JAK48271.1
KPJ20169.1 KQ459579 KPI99551.1 ODYU01005161 SOQ45754.1 NWSH01003475 PCG66221.1 KP657649 AKL78874.1 JTDY01001459 KOB73816.1 KQ971312 EEZ98455.2 NEVH01017443 PNF24483.1 PYGN01000762 PSN41105.1 KK852798 KDR16361.1 GEZM01075679 JAV64018.1 ATLV01019381 KE525269 KFB44107.1 AXCM01000067 ACPB03008308 CVRI01000065 CRL05790.1 LJIG01001596 KRT85344.1 GANO01001162 JAB58709.1 KK855354 PTY22144.1 CH477664 EAT37546.1 AAAB01008859 EAA07682.6 DS235866 EEB19367.1 JXUM01046136 KQ561464 KXJ78489.1 JRES01000062 KNC34475.1 GL439310 EFN67501.1 ADMH02000344 ETN66889.1 GL888056 EGI68598.1 NNAY01003307 OXU19647.1 AAZX01000328 KQ981522 KYN40548.1 JXJN01020792 GL453369 EFN76155.1 CCAG010012585 KQ760497 OAD60097.1 GL765283 EFZ16652.1 UFQS01001326 UFQT01001326 SSX10359.1 SSX30047.1 KQ414580 KOC71055.1 KQ976504 KYM82930.1 ADTU01023865 KQ979122 KYN22540.1 KZ288259 PBC30479.1 KK107238 QOIP01000007 EZA54672.1 RLU20763.1 JR035965 JR035966 JR035967 JR035968 AEY56830.1 AEY56831.1 AEY56832.1 AEY56833.1 DS231831 EDS31206.1 LBMM01000504 KMQ98193.1 GBYB01013930 JAG83697.1 AXCN02000061 GAMC01006914 JAB99641.1 CH964272 KRF99855.1 KQ434812 KZC06764.1 OUUW01000005 SPP80389.1 APCN01000427 CH933806 KRG02377.1 CP012526 ALC45474.1 CH917801 EDW04624.1 CH940656 EDW58394.1 CM000070 EIM52557.1 CH902617 KPU79602.1 GFXV01000249 MBW12054.1 BT120106 ADB13439.1 AF030197 AE014297 GGMR01005464 MBY18083.1 ABLF02025153 ABLF02025155 GGMS01008647 MBY77850.1 GL732531 EFX85953.1 ABJB011139869 DS910653 EEC16732.1 GDIP01071795 LRGB01001574 JAM31920.1 KZS11615.1 GDIQ01048214 JAN46523.1 LJIJ01000187 ODN00757.1 GDIP01010637 JAM93078.1 KY809750 AUN27676.1 LNIX01000063 OXA37206.1 GDIP01187824 JAJ35578.1 GDIP01187823 JAJ35579.1 GDIQ01207956 JAK43769.1 GDIQ01203454 JAK48271.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000037510
+ More
UP000007266 UP000235965 UP000245037 UP000076408 UP000027135 UP000075900 UP000030765 UP000075880 UP000075884 UP000075883 UP000015103 UP000183832 UP000075920 UP000075885 UP000075902 UP000008820 UP000007062 UP000076407 UP000075901 UP000009046 UP000069940 UP000249989 UP000037069 UP000075903 UP000000311 UP000069272 UP000000673 UP000007755 UP000079169 UP000215335 UP000002358 UP000078541 UP000092443 UP000092460 UP000078200 UP000008237 UP000092445 UP000092444 UP000091820 UP000053825 UP000078540 UP000005203 UP000005205 UP000192223 UP000095300 UP000078492 UP000242457 UP000095301 UP000053097 UP000279307 UP000075881 UP000002320 UP000036403 UP000075886 UP000007798 UP000076502 UP000192221 UP000268350 UP000075840 UP000009192 UP000092553 UP000001070 UP000008792 UP000001819 UP000007801 UP000000803 UP000007819 UP000000305 UP000001555 UP000076858 UP000094527 UP000198287
UP000007266 UP000235965 UP000245037 UP000076408 UP000027135 UP000075900 UP000030765 UP000075880 UP000075884 UP000075883 UP000015103 UP000183832 UP000075920 UP000075885 UP000075902 UP000008820 UP000007062 UP000076407 UP000075901 UP000009046 UP000069940 UP000249989 UP000037069 UP000075903 UP000000311 UP000069272 UP000000673 UP000007755 UP000079169 UP000215335 UP000002358 UP000078541 UP000092443 UP000092460 UP000078200 UP000008237 UP000092445 UP000092444 UP000091820 UP000053825 UP000078540 UP000005203 UP000005205 UP000192223 UP000095300 UP000078492 UP000242457 UP000095301 UP000053097 UP000279307 UP000075881 UP000002320 UP000036403 UP000075886 UP000007798 UP000076502 UP000192221 UP000268350 UP000075840 UP000009192 UP000092553 UP000001070 UP000008792 UP000001819 UP000007801 UP000000803 UP000007819 UP000000305 UP000001555 UP000076858 UP000094527 UP000198287
Interpro
SUPFAM
SSF103473
SSF103473
Gene 3D
CDD
ProteinModelPortal
H9JVA3
A0A212FCP6
A0A0N0PEL3
A0A194Q1Y0
A0A2H1VZT8
A0A2A4J2I7
+ More
A0A0G3VG09 A0A0L7LF40 D6W971 A0A2J7Q7C1 A0A2P8YA25 A0A182Y784 A0A067RCA1 A0A1Y1KTX3 A0A182RYG7 A0A084W1L3 A0A182J1B3 A0A182N4P3 A0A182M1V9 T1I654 A0A1J1J2W2 A0A182W2X5 A0A0T6BDE1 U5EX12 A0A182PA41 A0A182UE01 A0A2R7WRG2 A0A182UGY9 Q16SX3 Q7PR83 A0A182XIB7 A0A182T7R7 E0W161 A0A182G771 A0A0L0CQ39 A0A182VCH4 E2AGG8 A0A182F6G2 W5JVM7 F4WB43 A0A1S3D026 A0A232EMV3 K7J534 A0A195FJG4 A0A1A9XZF5 A0A1B0BUL9 A0A1A9VIY3 E2C7G8 A0A1A9ZL63 A0A1B0GET2 A0A310SPT4 A0A1A9W5V3 E9IS27 A0A336MP64 A0A0L7RJK5 A0A195BFI5 A0A087ZWQ0 A0A158NRC4 A0A1W4X2P3 A0A1I8NUP2 A0A151JBS7 A0A2A3EGY5 A0A1I8M3G4 A0A026WF51 V9I6X8 A0A182KBJ2 B0W363 A0A0J7L6J2 A0A0C9RXM3 A0A182QP81 W8C1Q4 A0A0Q9WUM0 A0A154P5Y7 A0A1W4UPF5 A0A3B0K361 A0A182HTA2 A0A0Q9X4V4 A0A0M5J8U6 B4K1S6 B4MBC3 I5ANM2 A0A0P8XY20 A0A2H8TD87 D2NUG3 O17444 A0A2S2NLL7 J9JXZ7 A0A2S2QJZ0 E9G3N1 B7QD10 A0A0P5XI13 A0A0P6G6T8 A0A1D2N677 A0A0P6C1V8 A0A2I6QBP8 A0A226CYU2 A0A0P5B8W8 A0A0P5BAW4 A0A0P5IXP8 A0A0P5KY64
A0A0G3VG09 A0A0L7LF40 D6W971 A0A2J7Q7C1 A0A2P8YA25 A0A182Y784 A0A067RCA1 A0A1Y1KTX3 A0A182RYG7 A0A084W1L3 A0A182J1B3 A0A182N4P3 A0A182M1V9 T1I654 A0A1J1J2W2 A0A182W2X5 A0A0T6BDE1 U5EX12 A0A182PA41 A0A182UE01 A0A2R7WRG2 A0A182UGY9 Q16SX3 Q7PR83 A0A182XIB7 A0A182T7R7 E0W161 A0A182G771 A0A0L0CQ39 A0A182VCH4 E2AGG8 A0A182F6G2 W5JVM7 F4WB43 A0A1S3D026 A0A232EMV3 K7J534 A0A195FJG4 A0A1A9XZF5 A0A1B0BUL9 A0A1A9VIY3 E2C7G8 A0A1A9ZL63 A0A1B0GET2 A0A310SPT4 A0A1A9W5V3 E9IS27 A0A336MP64 A0A0L7RJK5 A0A195BFI5 A0A087ZWQ0 A0A158NRC4 A0A1W4X2P3 A0A1I8NUP2 A0A151JBS7 A0A2A3EGY5 A0A1I8M3G4 A0A026WF51 V9I6X8 A0A182KBJ2 B0W363 A0A0J7L6J2 A0A0C9RXM3 A0A182QP81 W8C1Q4 A0A0Q9WUM0 A0A154P5Y7 A0A1W4UPF5 A0A3B0K361 A0A182HTA2 A0A0Q9X4V4 A0A0M5J8U6 B4K1S6 B4MBC3 I5ANM2 A0A0P8XY20 A0A2H8TD87 D2NUG3 O17444 A0A2S2NLL7 J9JXZ7 A0A2S2QJZ0 E9G3N1 B7QD10 A0A0P5XI13 A0A0P6G6T8 A0A1D2N677 A0A0P6C1V8 A0A2I6QBP8 A0A226CYU2 A0A0P5B8W8 A0A0P5BAW4 A0A0P5IXP8 A0A0P5KY64
PDB
6GV1
E-value=0.010301,
Score=93
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
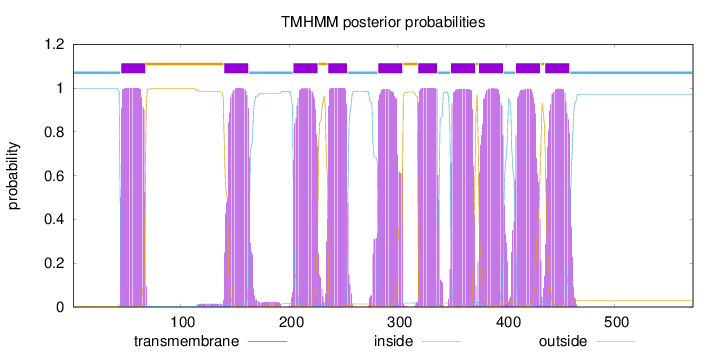
Length:
572
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
218.44818
Exp number, first 60 AAs:
16.40849
Total prob of N-in:
0.99698
POSSIBLE N-term signal
sequence
inside
1 - 44
TMhelix
45 - 67
outside
68 - 139
TMhelix
140 - 162
inside
163 - 203
TMhelix
204 - 226
outside
227 - 235
TMhelix
236 - 253
inside
254 - 281
TMhelix
282 - 304
outside
305 - 318
TMhelix
319 - 336
inside
337 - 348
TMhelix
349 - 371
outside
372 - 374
TMhelix
375 - 397
inside
398 - 408
TMhelix
409 - 431
outside
432 - 435
TMhelix
436 - 458
inside
459 - 572
Population Genetic Test Statistics
Pi
240.662422
Theta
176.219059
Tajima's D
1.198695
CLR
0.667057
CSRT
0.716014199290036
Interpretation
Uncertain
