Pre Gene Modal
BGIBMGA013442
Annotation
PREDICTED:_mitogen-activated_protein_kinase_kinase_kinase_4?_partial_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.763
Sequence
CDS
ATGGATATGCCAGACGGTGACTGGAAGAAGAAAGTTGGATTTCCGATCAATTACGACTCGAACCAGAGCTCCAGCGATGAGTTCGACTCATCAGATGTAGTGATGAGACATCCGCGCCAGTCGTCCAGGTACTTGCTCGCCGACGGCGGCGACGGAGCTGAAGCCGACCTCCTAGGTACGACGCCTCCACGCTCAAGAATCAGGGATAAAGAAAAGAGAAGAAGCACCATGGACCCCGGGCTGAATTTCCAACGGCTACTGCGAGACAAGAAGGACTCTGTTGAACTGGTACCTGAACCAATCCAGCCGCCTCTGGCACCAGAGCGCCTCAAGAAACGTGCCAGTCTGAAACTCATTCGGAACATGGAGAGGGATTGGAAACAAGACATCAATCAGAGAGAGAGCAAGGCGGACTCTCTTCCCTCGCCCAATCCAATCCCGACTGTTCCTACATTGGTAGAGTCGTGCAGCCGGTTCATGTCCCTGACGTCCCGGCCCAAGTGCCGCGAGATGATAGCTCAGGACGCGGCGGAAACGGAAGTGGAGGAGCGCTGGCGATCGGACCGCGCAGACGAAGTGCCCCCCAGCCGAGTCAACTTCCACAAGACGTTCAGTATGCTAATCAACATGGGGAATATCGAGAAAGGGTGCAGAAGGACTATCAGCCGCGAAGAACAAGTTTGGCAAAACGAGCTGAAGGATCTGATCTGGTTGGAGCTGCAGGCCAAGATCGCCGGGCGGACCCTCGCCGACCAGGACCAGTACCTGTGCGCCCAAAGAGACATCGTGCCAACTCTCATCAAGAACATTTTTGAGTATAGGTTCGTAGACAATAACCCGTGTAAGGACCAGTCGAAGAAGCTGACGGAAGCTGTGAACGCCGATGTCAGTGTCGAACATGTCGCCGGAGAGGAGTATGATGACGAGGGCTGCGCGCTGGGCTGCGCGTCGCTGGAGTGCGAGACGTGCTCTGGGCGCGTGGGCGGCGCCATGCGCGAGGTGGGCACGCTGCTCGACTCCTTCTACAACGCGCACGCGCTCTATCCCTCCAGCAAGGCCATGACGGCGGAGCACCCGCTCATAGCCACCAACCTCTTCAAGAACCGGATCAAGGCGATGTGTCTCTGGTACAACACGGCGCTGAACATGCGCGTGAAGCTGCTGGCGGTGCGCCGCATGCTGCGGTCCATGCTCCTCAAGCGCCGCCCCGCCCCGCACCGCAGTTCCGTGGACTCCGACGTCAGCAGGCAGTCTTCGGAGATCCAAAGGCAGTGCCAGGTCAGGTTCAACGTCAGTGTCCACGCGTCAGACTCCAACAGCAGCGACAGCAGCGGCCACTCGGAGATAAAGGAAGAACCGAAAACAGAAGTGAGGAGTGAGGGCAGCAGCGAGTCCCCCGCGGGCGCGCACCGGCGGGGGGGCGGGGGCGAGGGGGACCCCTCCAAGCTGAACGGCCAAGTGACCCCCGAGATAGTGATCTCTGAGAGCCAGTACAGCACGGGCGAGGAGAGCAGCTACTCCACGGAGAGCTGGTCGTCGTGGTGGTCCAGCCTCGACCCGCTGGAGCTGGGCCCGCTGTCCGAGCTGACGCAGCTGCGGCTCCTCGACAAGTGCCCCGTGTCCCTCTACCGGGAGCATCACCGCGAGATCCTGAAGACGCAGGGCGTTCGTCGCAGCATGATGTTCATATACAAGATGAAGAACAACTTGCTGCACAAAGTACACTTGACCCTGGAGAAGCCTGACGCCACCACTGACACGTTCGCGGAGCACGAGCCCGTCGAGGTGTTTGAAGACTCGAAGAACGAAGACAAGTCGGAATCGATGCAGGACGAGAACGACAAGAGAAACTACGAGCTCAGAAGATACGGCTGGTGGTCCGAAGAGACGTCAGCGATGCGCCTGCCTTCCTACCGAGGACATTTCCTGCTGCTGTCCAGCATCTGTCTCGAAGCCGTACACGACTACCTCTCTTTCAGGCTCGAGGCTCGACCGGACCGGCCCTCTTGCCTCACCGTTAAACAGCTCATCCACGAGCTGAAGGAGGGTCTGGACATAGCGTCCGAGATGCGCTTTGACTTCGTGCGCAACGTGCGCGCGGCGCTCGCGGGCCGGAGCCCCGGGACAGCGGCAACTCAGCTACGTCTGATGACCAACACGTACGACGACACCGTGAGGGATGTGCTCAAGCAATACCTGTCGTACCTGACACTGATGTCGGAGACGGAGCACATGTCCCGCGCGGGGCTGCAGGCCGAGTGGGAGTACACGGCGCGGCTCGCCCGCCGCATCCACTGCGCCTCGCAGCTGGCGCCGCTCGCCTTCTCTGAAATAGCTTGCAACCAGTTGAATAGGCTGTTGAAAAGCTTCGAAGATAACTTTCAACAGCTAATTCAGTCGGTGCATGCTGACATCGAAACTAACAGACACGCGGTGTACGCCGTGTGCCGGCTCGCGCAGAGCATGTACACGTGCGAGCGGGAGCGCACGCTGCAGGCGGCGCAGTGGGCGCGCGCACTCAGCGCCCGGCTGCAGCGCGCGCCGCCGCCCGCCTACATGCCGCAGCGCGCCCTCATCTTCGAAAGCCTCATGAAAATACGCGACTTCATAGTGAAAAACGTCGAGTACATACTGGAGCAGAGCAAGACGACGCCGGACGACCTCACCAAGGAGATGCACGAGAACGTGGTGGCCAGGATCAGGGAGCTCCTGCTGCAGATATTCAAATTAGGATTCGAAATCCACAAGGAGCTGCACCGCTTCGCGAACGATTCGACGCGGCTATCGTTGCCCCCCCGCCGGTCGGTGGGCGGCGCGCCCTCCCGGTACCGGAGGAGGGTGCTCAGGACTGACAGCGTCTTGGAGGACGGAGAAGCCAGCGTTTTTGATGACAGACTCTTTGACAACCAGTTCGGGGAGAATCAGACTCAAACGAGCATCGAACTACTACTATCCGGTGAGCGAGCGTCAGATCCTCCGGTGGAGGCTGCGGAGGGGGGAGGAGGGGAGGCGGTCGTCGAGGGGGAGGAGGTAGAGCGGAGGTGGGCGGGGCGGGTGGCGCGAGCTGTGATACAGTTCGCGACCTGCTGGATGCAGTTCGTGCTGGAGCGCTGCGACCGCGGCAGAGGCACCAGACCTAGATGGGCGAGTCAAGGGCTCGAGTTTCTGATGTTAGCCTGTGATCCTTGCAACACTCAATATTTGACTGATGAAGAATTTGAGGAGCTGAAGCAGCGCATGGACAGCTGCATCTCGCACGTGATCGGGTCCCGCAGCGCGCGCACGCCCGACCCCGCGCCCCTCGCCTCGCCGCGCCCCCGCGCCCGCCTGCACGTACGCACCCCCATTAACTCCGCCCAACAATTAATCTTGTCACTTCAAGATCCAAAATTACACGTTGCTAATATTTTGTGCATTGAATCTAATTTGGGGAGTTTGTACGAAGGAACTCTCCGAATCCTGATCACGGAGCCAGTTACCGTCGACGCCCTAGACACTTGCTTACGGATCCATTAG
Protein
MDMPDGDWKKKVGFPINYDSNQSSSDEFDSSDVVMRHPRQSSRYLLADGGDGAEADLLGTTPPRSRIRDKEKRRSTMDPGLNFQRLLRDKKDSVELVPEPIQPPLAPERLKKRASLKLIRNMERDWKQDINQRESKADSLPSPNPIPTVPTLVESCSRFMSLTSRPKCREMIAQDAAETEVEERWRSDRADEVPPSRVNFHKTFSMLINMGNIEKGCRRTISREEQVWQNELKDLIWLELQAKIAGRTLADQDQYLCAQRDIVPTLIKNIFEYRFVDNNPCKDQSKKLTEAVNADVSVEHVAGEEYDDEGCALGCASLECETCSGRVGGAMREVGTLLDSFYNAHALYPSSKAMTAEHPLIATNLFKNRIKAMCLWYNTALNMRVKLLAVRRMLRSMLLKRRPAPHRSSVDSDVSRQSSEIQRQCQVRFNVSVHASDSNSSDSSGHSEIKEEPKTEVRSEGSSESPAGAHRRGGGGEGDPSKLNGQVTPEIVISESQYSTGEESSYSTESWSSWWSSLDPLELGPLSELTQLRLLDKCPVSLYREHHREILKTQGVRRSMMFIYKMKNNLLHKVHLTLEKPDATTDTFAEHEPVEVFEDSKNEDKSESMQDENDKRNYELRRYGWWSEETSAMRLPSYRGHFLLLSSICLEAVHDYLSFRLEARPDRPSCLTVKQLIHELKEGLDIASEMRFDFVRNVRAALAGRSPGTAATQLRLMTNTYDDTVRDVLKQYLSYLTLMSETEHMSRAGLQAEWEYTARLARRIHCASQLAPLAFSEIACNQLNRLLKSFEDNFQQLIQSVHADIETNRHAVYAVCRLAQSMYTCERERTLQAAQWARALSARLQRAPPPAYMPQRALIFESLMKIRDFIVKNVEYILEQSKTTPDDLTKEMHENVVARIRELLLQIFKLGFEIHKELHRFANDSTRLSLPPRRSVGGAPSRYRRRVLRTDSVLEDGEASVFDDRLFDNQFGENQTQTSIELLLSGERASDPPVEAAEGGGGEAVVEGEEVERRWAGRVARAVIQFATCWMQFVLERCDRGRGTRPRWASQGLEFLMLACDPCNTQYLTDEEFEELKQRMDSCISHVIGSRSARTPDPAPLASPRPRARLHVRTPINSAQQLILSLQDPKLHVANILCIESNLGSLYEGTLRILITEPVTVDALDTCLRIH
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
Ontologies
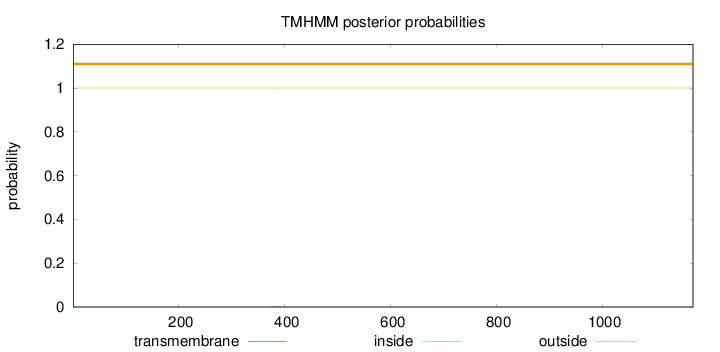
Topology
Length:
1171
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00711999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00023
outside
1 - 1171
Population Genetic Test Statistics
Pi
227.677909
Theta
180.918289
Tajima's D
0.621708
CLR
0.043642
CSRT
0.550722463876806
Interpretation
Uncertain
