Gene
KWMTBOMO03153
Pre Gene Modal
BGIBMGA013436
Annotation
PREDICTED:_scavenger_receptor_class_B_member_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.014
Sequence
CDS
ATGTTCTTCGATCCAGTAATACCTATTACATTATTTTATTCCCGAGCAGCCAATGGTACGCTTACTCACAACGCCATGATGGAAGTTATAGAAGGTATACACATATCGGCGTACCTGTTCAACATTACCAACGCTGAGGAGTTCACGTCCGGAGAGCATGATAAGCTGAAAGTGCAAGAAGTCGGGCCGTTTACTTATAGTGAGGCTCGAACGAATAAAAACTTTGTGATAGATGAAGAAGCTGAAGAAATGAGGTATACTCCTCGAATCGATGTGAACTTCATACCCGAGGCGTCTGTGGGAAGGCCTGAAGATATAGATGTCACGTTGGCGAATATCGCAATGTTGAGCATAGCCTCCAAAGTGGGTGCCTACTCCTACTTTCCTCAACTAGCCTTCAACCTTCTGGCATCGCAGCTGAACTCCAAGCCAGTAGTGACGATGAGAGCAAAGGACTACCTCTGGGGCTACGACGAGCCTCTTATAAGATTAGGGCATAATATTATACCAGGGATAATAAACTTCGATAGATTGGGAATTTTGGATAGACTTTACGATAAAAAAGCGGTCTATAACTATGTGGTGAGCGCTAAAAACTCAGACAAGTTTGCCATTAAGTCTGTAAATGGAATTGAAGGTCTTGAAATGTGGAATTACGAAAATCCCAAGAAAAGGTCCAAATGCAACTCATTTATAGACGCATTTGAAGGTATCAACTACCCCGCCATGATGACCCCAGAAACGCCTATAAGAATATACAGAAGTATTCTCTGTCGAATTCTGGAATTGGATTATTTTGACACGATAACAACTGACTATGGAGCCAAGGGTCTGGTCTATCGAATTAGTAACAGAACGTATACGACAAATAACGATACCAAATGTCTTTGTATTGGAGAATGTCAAGAGTACATTTCTCATCTGTCACCCTGCTTTTTTGGTCTACCCGTGGCTCTATCTAATGCACATTTCCTGTACGCCGATCCAGAAATTTATAGCAAAATAGAAGGAATTCAACCTGACGAAGAGAAACATGGCAGCAATATACTAATTGACCCGATAATGGGCTTGGCGTTGAAAACCGAATTTTCAGTACAACTAAACGTAGTGGTTGATGATGTGACTTTCAACAGTGACGCTAGAAGATTCTCAAAAATGCTGGTACCAGTGGGGTATTTCAAAATTGTACAGCCACCTCTTAAAAGCACTCTGATATTTCAACTGAGATTGGTATATGTCTATGGACCATATTTGGTGTTAGCAGTACAAATTATTTTCTTGGTTCTCGGCTTAACAATATTGGCTTATTCTCGTCGGAACATGAATTGCGGATTACTTTATCTCACACGAAAGGAAATTACTTTTTATTTGAGCACAGCGGAGAAAGCTAAAGTCGAACAGCCATTGATTTATTGA
Protein
MFFDPVIPITLFYSRAANGTLTHNAMMEVIEGIHISAYLFNITNAEEFTSGEHDKLKVQEVGPFTYSEARTNKNFVIDEEAEEMRYTPRIDVNFIPEASVGRPEDIDVTLANIAMLSIASKVGAYSYFPQLAFNLLASQLNSKPVVTMRAKDYLWGYDEPLIRLGHNIIPGIINFDRLGILDRLYDKKAVYNYVVSAKNSDKFAIKSVNGIEGLEMWNYENPKKRSKCNSFIDAFEGINYPAMMTPETPIRIYRSILCRILELDYFDTITTDYGAKGLVYRISNRTYTTNNDTKCLCIGECQEYISHLSPCFFGLPVALSNAHFLYADPEIYSKIEGIQPDEEKHGSNILIDPIMGLALKTEFSVQLNVVVDDVTFNSDARRFSKMLVPVGYFKIVQPPLKSTLIFQLRLVYVYGPYLVLAVQIIFLVLGLTILAYSRRNMNCGLLYLTRKEITFYLSTAEKAKVEQPLIY
Summary
Similarity
Belongs to the CD36 family.
Uniprot
A0A2W1BT84
A0A2A4JT04
A0A2H1V8Y8
A0A0L7KUM6
A0A194Q1W4
A0A212FCS4
+ More
A0A2H1V5V1 A0A0N0PEN1 K7ZN68 K7ZLU1 K7ZRZ1 H9JV76 A0A2A4JTS4 K7ZQ90 K7ZLU0 A0A2H1V7C0 H9JV74 A0A2W1BRR6 A0A034VI50 A0A0K8WGU0 A0A0L0BYA6 T1HPT0 A0A0A1XC80 A0A023F2D4 E2BXL2 E0W134 A0A0N0U3K8 A0A2A3EK48 A0A1B6CDS3 A0A194Q3D1 A0A2S2QNL3 A0A151WH40 D6W974 A0A195E3W4 F4WJM4 T1PIR5 A0A2H8TL09 A0A088AD42 A0A0L7QWT1 A0A1B0FEG9 A0A026X1F9 K7ISQ1 J9KAS8 A0A1W4WGT2 B4JU08 A0A1A9ZZ29 A0A195FW92 A0A1W4UIZ8 A0A195AV35 A0A1A9VFW7 A0A0A9YUX7 A0A151I684 A0A154PGJ9 A0A3B0K495 A0A3B0JEC8 H9JVK9 A0A1I8NTR6 B3NZD7 A0A1Y1MG67 B4PMY6 A0A2J7PW76 E2AWE0 A0A336L8K1 A0A1A9WGN3 A0A2W1BDC0 A0A0N8P1E2 A0A1I8N7C1 B3M2M0 B4LXR0 B4IBT3 A0A1I8N7B3 B4QWH0 U4U1T1 A0A336LYF7 B4K5G4 N6UAS4 A0A1I8N7B6 A0A2S2Q087 A0A232F2G1 B4NF45 N0A138 A0A232EZ92 B4GZ30 Q9Y121 K7IRA8 Q9VET1 A0A1A9YSY3 Q29B54 A0A310SMC7 A0A1B0B2M1 A0A139WG73 A0A1L8DJL5 A0A1L8DJK2 A0A1L8DJY4 A0A067QYX6 Q7Q7R9 A0A0L7QQ97 A0A0L0C5C4 J9JX78
A0A2H1V5V1 A0A0N0PEN1 K7ZN68 K7ZLU1 K7ZRZ1 H9JV76 A0A2A4JTS4 K7ZQ90 K7ZLU0 A0A2H1V7C0 H9JV74 A0A2W1BRR6 A0A034VI50 A0A0K8WGU0 A0A0L0BYA6 T1HPT0 A0A0A1XC80 A0A023F2D4 E2BXL2 E0W134 A0A0N0U3K8 A0A2A3EK48 A0A1B6CDS3 A0A194Q3D1 A0A2S2QNL3 A0A151WH40 D6W974 A0A195E3W4 F4WJM4 T1PIR5 A0A2H8TL09 A0A088AD42 A0A0L7QWT1 A0A1B0FEG9 A0A026X1F9 K7ISQ1 J9KAS8 A0A1W4WGT2 B4JU08 A0A1A9ZZ29 A0A195FW92 A0A1W4UIZ8 A0A195AV35 A0A1A9VFW7 A0A0A9YUX7 A0A151I684 A0A154PGJ9 A0A3B0K495 A0A3B0JEC8 H9JVK9 A0A1I8NTR6 B3NZD7 A0A1Y1MG67 B4PMY6 A0A2J7PW76 E2AWE0 A0A336L8K1 A0A1A9WGN3 A0A2W1BDC0 A0A0N8P1E2 A0A1I8N7C1 B3M2M0 B4LXR0 B4IBT3 A0A1I8N7B3 B4QWH0 U4U1T1 A0A336LYF7 B4K5G4 N6UAS4 A0A1I8N7B6 A0A2S2Q087 A0A232F2G1 B4NF45 N0A138 A0A232EZ92 B4GZ30 Q9Y121 K7IRA8 Q9VET1 A0A1A9YSY3 Q29B54 A0A310SMC7 A0A1B0B2M1 A0A139WG73 A0A1L8DJL5 A0A1L8DJK2 A0A1L8DJY4 A0A067QYX6 Q7Q7R9 A0A0L7QQ97 A0A0L0C5C4 J9JX78
Pubmed
28756777
26227816
26354079
22118469
23160179
19121390
+ More
25348373 26108605 25830018 25474469 20798317 20566863 18362917 19820115 21719571 25315136 24508170 30249741 20075255 17994087 25401762 28004739 17550304 23537049 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 24845553 12364791
25348373 26108605 25830018 25474469 20798317 20566863 18362917 19820115 21719571 25315136 24508170 30249741 20075255 17994087 25401762 28004739 17550304 23537049 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 24845553 12364791
EMBL
KZ149979
PZC75846.1
NWSH01000627
PCG75175.1
ODYU01001113
SOQ36872.1
+ More
JTDY01005621 KOB66820.1 KQ459579 KPI99536.1 AGBW02009155 OWR51508.1 ODYU01000809 SOQ36156.1 KQ459753 KPJ20156.1 AB742536 AB742537 BAM67015.1 BAM67016.1 AB742541 BAM67018.1 AB742538 BAM67017.1 BABH01037377 BABH01037378 PCG75176.1 AB742535 BAM67014.1 AB742534 BAM67013.1 SOQ36154.1 BABH01037379 BABH01037380 BABH01037381 PZC75847.1 GAKP01016788 JAC42164.1 GDHF01001943 JAI50371.1 JRES01001160 KNC24951.1 ACPB03008273 GBXI01005740 JAD08552.1 GBBI01003543 JAC15169.1 GL451265 EFN79581.1 DS235866 EEB19340.1 KQ435878 KOX69994.1 KZ288220 PBC32145.1 GEDC01025692 JAS11606.1 KPI99519.1 GGMS01010060 MBY79263.1 KQ983136 KYQ47151.1 KQ971312 EEZ99259.1 KQ979685 KYN19838.1 GL888186 EGI65516.1 KA647995 AFP62624.1 GFXV01003022 MBW14827.1 KQ414706 KOC63072.1 CCAG010014864 KK107031 QOIP01000008 EZA62097.1 RLU19322.1 AAZX01007991 ABLF02039108 CH916374 EDV91587.1 KQ981208 KYN44713.1 KQ976738 KYM75834.1 GBHO01010229 JAG33375.1 KQ978495 KYM93627.1 KQ434889 KZC10300.1 OUUW01000005 SPP80456.1 SPP80455.1 BABH01032481 BABH01032482 BABH01032483 CH954181 EDV48809.1 GEZM01032287 GEZM01032286 JAV84694.1 CM000160 EDW96998.1 NEVH01020937 PNF20584.1 GL443286 EFN62240.1 UFQS01002145 UFQT01002145 SSX13321.1 SSX32757.1 KZ150255 PZC71794.1 CH902617 KPU79676.1 EDV42341.1 CH940650 EDW67869.1 CH480827 EDW44841.1 CM000364 EDX12661.1 KB631903 ERL87022.1 UFQT01000186 SSX21297.1 CH933806 EDW16190.1 APGK01042140 APGK01042141 KB741002 ENN75732.1 GGMS01001943 MBY71146.1 NNAY01001156 OXU24944.1 CH964251 EDW83420.2 BT150082 AGK43573.1 NNAY01001554 OXU23617.1 CH479198 EDW28048.1 AF145651 AAD38626.1 AE014297 BT044340 AAF55338.1 ACH92405.1 CM000070 EAL27145.1 KQ762196 OAD56141.1 JXJN01007683 KQ971348 KYB26889.1 GFDF01007426 JAV06658.1 GFDF01007425 JAV06659.1 GFDF01007424 JAV06660.1 KK853094 KDR11500.1 AAAB01008952 EAA10557.4 KQ414792 KOC60729.1 JRES01000973 KNC26639.1 ABLF02026897 ABLF02026902
JTDY01005621 KOB66820.1 KQ459579 KPI99536.1 AGBW02009155 OWR51508.1 ODYU01000809 SOQ36156.1 KQ459753 KPJ20156.1 AB742536 AB742537 BAM67015.1 BAM67016.1 AB742541 BAM67018.1 AB742538 BAM67017.1 BABH01037377 BABH01037378 PCG75176.1 AB742535 BAM67014.1 AB742534 BAM67013.1 SOQ36154.1 BABH01037379 BABH01037380 BABH01037381 PZC75847.1 GAKP01016788 JAC42164.1 GDHF01001943 JAI50371.1 JRES01001160 KNC24951.1 ACPB03008273 GBXI01005740 JAD08552.1 GBBI01003543 JAC15169.1 GL451265 EFN79581.1 DS235866 EEB19340.1 KQ435878 KOX69994.1 KZ288220 PBC32145.1 GEDC01025692 JAS11606.1 KPI99519.1 GGMS01010060 MBY79263.1 KQ983136 KYQ47151.1 KQ971312 EEZ99259.1 KQ979685 KYN19838.1 GL888186 EGI65516.1 KA647995 AFP62624.1 GFXV01003022 MBW14827.1 KQ414706 KOC63072.1 CCAG010014864 KK107031 QOIP01000008 EZA62097.1 RLU19322.1 AAZX01007991 ABLF02039108 CH916374 EDV91587.1 KQ981208 KYN44713.1 KQ976738 KYM75834.1 GBHO01010229 JAG33375.1 KQ978495 KYM93627.1 KQ434889 KZC10300.1 OUUW01000005 SPP80456.1 SPP80455.1 BABH01032481 BABH01032482 BABH01032483 CH954181 EDV48809.1 GEZM01032287 GEZM01032286 JAV84694.1 CM000160 EDW96998.1 NEVH01020937 PNF20584.1 GL443286 EFN62240.1 UFQS01002145 UFQT01002145 SSX13321.1 SSX32757.1 KZ150255 PZC71794.1 CH902617 KPU79676.1 EDV42341.1 CH940650 EDW67869.1 CH480827 EDW44841.1 CM000364 EDX12661.1 KB631903 ERL87022.1 UFQT01000186 SSX21297.1 CH933806 EDW16190.1 APGK01042140 APGK01042141 KB741002 ENN75732.1 GGMS01001943 MBY71146.1 NNAY01001156 OXU24944.1 CH964251 EDW83420.2 BT150082 AGK43573.1 NNAY01001554 OXU23617.1 CH479198 EDW28048.1 AF145651 AAD38626.1 AE014297 BT044340 AAF55338.1 ACH92405.1 CM000070 EAL27145.1 KQ762196 OAD56141.1 JXJN01007683 KQ971348 KYB26889.1 GFDF01007426 JAV06658.1 GFDF01007425 JAV06659.1 GFDF01007424 JAV06660.1 KK853094 KDR11500.1 AAAB01008952 EAA10557.4 KQ414792 KOC60729.1 JRES01000973 KNC26639.1 ABLF02026897 ABLF02026902
Proteomes
UP000218220
UP000037510
UP000053268
UP000007151
UP000053240
UP000005204
+ More
UP000037069 UP000015103 UP000008237 UP000009046 UP000053105 UP000242457 UP000075809 UP000007266 UP000078492 UP000007755 UP000095301 UP000005203 UP000053825 UP000092444 UP000053097 UP000279307 UP000002358 UP000007819 UP000192223 UP000001070 UP000092445 UP000078541 UP000192221 UP000078540 UP000078200 UP000078542 UP000076502 UP000268350 UP000095300 UP000008711 UP000002282 UP000235965 UP000000311 UP000091820 UP000007801 UP000008792 UP000001292 UP000000304 UP000030742 UP000009192 UP000019118 UP000215335 UP000007798 UP000008744 UP000000803 UP000092443 UP000001819 UP000092460 UP000027135 UP000007062
UP000037069 UP000015103 UP000008237 UP000009046 UP000053105 UP000242457 UP000075809 UP000007266 UP000078492 UP000007755 UP000095301 UP000005203 UP000053825 UP000092444 UP000053097 UP000279307 UP000002358 UP000007819 UP000192223 UP000001070 UP000092445 UP000078541 UP000192221 UP000078540 UP000078200 UP000078542 UP000076502 UP000268350 UP000095300 UP000008711 UP000002282 UP000235965 UP000000311 UP000091820 UP000007801 UP000008792 UP000001292 UP000000304 UP000030742 UP000009192 UP000019118 UP000215335 UP000007798 UP000008744 UP000000803 UP000092443 UP000001819 UP000092460 UP000027135 UP000007062
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BT84
A0A2A4JT04
A0A2H1V8Y8
A0A0L7KUM6
A0A194Q1W4
A0A212FCS4
+ More
A0A2H1V5V1 A0A0N0PEN1 K7ZN68 K7ZLU1 K7ZRZ1 H9JV76 A0A2A4JTS4 K7ZQ90 K7ZLU0 A0A2H1V7C0 H9JV74 A0A2W1BRR6 A0A034VI50 A0A0K8WGU0 A0A0L0BYA6 T1HPT0 A0A0A1XC80 A0A023F2D4 E2BXL2 E0W134 A0A0N0U3K8 A0A2A3EK48 A0A1B6CDS3 A0A194Q3D1 A0A2S2QNL3 A0A151WH40 D6W974 A0A195E3W4 F4WJM4 T1PIR5 A0A2H8TL09 A0A088AD42 A0A0L7QWT1 A0A1B0FEG9 A0A026X1F9 K7ISQ1 J9KAS8 A0A1W4WGT2 B4JU08 A0A1A9ZZ29 A0A195FW92 A0A1W4UIZ8 A0A195AV35 A0A1A9VFW7 A0A0A9YUX7 A0A151I684 A0A154PGJ9 A0A3B0K495 A0A3B0JEC8 H9JVK9 A0A1I8NTR6 B3NZD7 A0A1Y1MG67 B4PMY6 A0A2J7PW76 E2AWE0 A0A336L8K1 A0A1A9WGN3 A0A2W1BDC0 A0A0N8P1E2 A0A1I8N7C1 B3M2M0 B4LXR0 B4IBT3 A0A1I8N7B3 B4QWH0 U4U1T1 A0A336LYF7 B4K5G4 N6UAS4 A0A1I8N7B6 A0A2S2Q087 A0A232F2G1 B4NF45 N0A138 A0A232EZ92 B4GZ30 Q9Y121 K7IRA8 Q9VET1 A0A1A9YSY3 Q29B54 A0A310SMC7 A0A1B0B2M1 A0A139WG73 A0A1L8DJL5 A0A1L8DJK2 A0A1L8DJY4 A0A067QYX6 Q7Q7R9 A0A0L7QQ97 A0A0L0C5C4 J9JX78
A0A2H1V5V1 A0A0N0PEN1 K7ZN68 K7ZLU1 K7ZRZ1 H9JV76 A0A2A4JTS4 K7ZQ90 K7ZLU0 A0A2H1V7C0 H9JV74 A0A2W1BRR6 A0A034VI50 A0A0K8WGU0 A0A0L0BYA6 T1HPT0 A0A0A1XC80 A0A023F2D4 E2BXL2 E0W134 A0A0N0U3K8 A0A2A3EK48 A0A1B6CDS3 A0A194Q3D1 A0A2S2QNL3 A0A151WH40 D6W974 A0A195E3W4 F4WJM4 T1PIR5 A0A2H8TL09 A0A088AD42 A0A0L7QWT1 A0A1B0FEG9 A0A026X1F9 K7ISQ1 J9KAS8 A0A1W4WGT2 B4JU08 A0A1A9ZZ29 A0A195FW92 A0A1W4UIZ8 A0A195AV35 A0A1A9VFW7 A0A0A9YUX7 A0A151I684 A0A154PGJ9 A0A3B0K495 A0A3B0JEC8 H9JVK9 A0A1I8NTR6 B3NZD7 A0A1Y1MG67 B4PMY6 A0A2J7PW76 E2AWE0 A0A336L8K1 A0A1A9WGN3 A0A2W1BDC0 A0A0N8P1E2 A0A1I8N7C1 B3M2M0 B4LXR0 B4IBT3 A0A1I8N7B3 B4QWH0 U4U1T1 A0A336LYF7 B4K5G4 N6UAS4 A0A1I8N7B6 A0A2S2Q087 A0A232F2G1 B4NF45 N0A138 A0A232EZ92 B4GZ30 Q9Y121 K7IRA8 Q9VET1 A0A1A9YSY3 Q29B54 A0A310SMC7 A0A1B0B2M1 A0A139WG73 A0A1L8DJL5 A0A1L8DJK2 A0A1L8DJY4 A0A067QYX6 Q7Q7R9 A0A0L7QQ97 A0A0L0C5C4 J9JX78
PDB
5UPH
E-value=8.70189e-25,
Score=282
Ontologies
GO
PANTHER
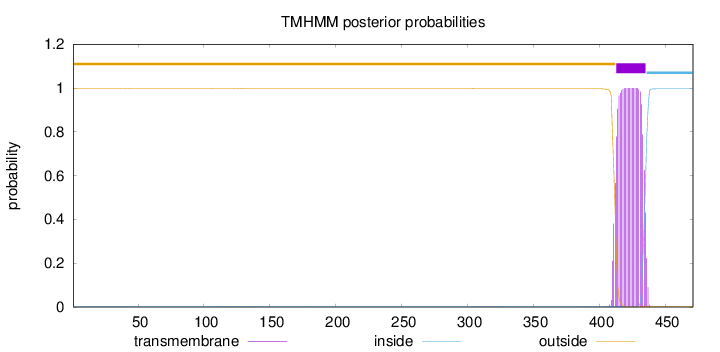
Topology
Length:
471
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.99619
Exp number, first 60 AAs:
0.01815
Total prob of N-in:
0.00294
outside
1 - 412
TMhelix
413 - 435
inside
436 - 471
Population Genetic Test Statistics
Pi
213.29256
Theta
163.100338
Tajima's D
0.591437
CLR
0.485424
CSRT
0.543972801359932
Interpretation
Uncertain
