Gene
KWMTBOMO03148
Pre Gene Modal
BGIBMGA013431
Annotation
PREDICTED:_hemicentin-2-like_[Bombyx_mori]
Full name
Protogenin
Alternative Name
Protein Shen-Dan
Location in the cell
Cytoplasmic Reliability : 1.114
Sequence
CDS
ATGAAGAAGTTCAAAAATTTACCAGTGACATGGGCTCCCGCGCGCACGCTACTGCCGTGCGTGGTACCCGACGGAGAGACGCTGGTGGTGCGCGGCCCGCCACTACTCCTCGACTCCGATGACCAGAACCGTCTCAAGCCTTACGTTTGGATTAAGGATAACAAGGAGATTAAGCAGTCGTTAACGAACGTGACCCTAGAGGTACTGCAGAAGAAGGATGGCACCAGCGAGGGGGTCTACCAGTGCGGGGTGAGGCACCATGGCCTGGTGTTGGGCTACCCCATTATTCTCAAGTTTGCCTTCATCGAAAAGTACGTCTCTGTGGTTCCTGAGAACAGCACCGTGCGCGCCGGGCAGCCGTTTGTGCTCACGTGCAACATCAGCTCGGGTCCGTCTGCCAGTATCACGTGGCTGAAAGGAGAGGAAGTCATATCAGAGAACGACAGATATTATTACATCGACAATAAGCATCAACTACTAATACTGAACGCGAAGGAACAAGATGCAGGCGTATATTGGTGTAAAGCAACCAACCCAGTGCTAAACAAGAGTAGAAACAGTAGTCCTGCTTTGGTACAGGTGGAGAGGGAGGAAGGTCAGACGGGGCTGGCCTTTCTTCCTCTGCAGCAAGACACCACTATCTACGTCTCGAAGGGGAGCAGAGTTGTACTCCCATGTCCAGTAGTGGGCTGGCCCAAGCCCGAGATTGTATGGAAGCTGACTCCATCGATCGGAAGAACCGCTGAGCAAGAGACTATAGACCAGGTGTTGGTACTGCCGAGCATGCAGGCGGATCTAGAGGGAGTCTTCTCGTGCTCAGTGGACGGGAATAATGATTTGGTTAAGACCTACAATGTTACGATTACGGAGCCAGTGAACATCACGTTGCCTCCGGCTTCAAAGCAGGTGTACCGGGCCAGCACGGTGAGATTCAACTGCACCGCCACCGGGAAACCGGCCCCGATCATCACCTGGTACAAGGACGGGAAGCCTCTGGTTCTGGCCGGCAGGAAAGTTGTACTGACTTCGTTTGAAGGGAATCGTCGAGAGCTACTCATTAGGAGTGTTACATCTGATGATGCTGGAGTGTACCAGTGCTTCGCTAGGAACGCGCTGCAGGAGCGCGGCGCGTGGGCGCGGCTGGAGGTGGCGGGCGCGGGGGCGGAGGCGCCGCGCGGGGTGCGGTGTGCGGCGGCGGGGCCCGCGCGGGTGCGCGTGTACTGGGACCTGATGGACGCCGACGTTGTGGCCTACACCGTGCAGCGGAGCGTGTCGCAAGGTACCATGTACCCTGGAGTGCCGCGAACTACCAACGAAGAAATCATCACCGTAAATGAACCATTGACTCCTTACAAATTTCAGGTGACGGCTTATATTCTGACCAAAAACAATAAGACCATAGCGAGCGATCCCTCAGAAACTGTCACCTGTCAGGGGCAGGGAGTGCCGATCAAGATAACGCGAGTGAACGACGAGAGGGTCGTGGTATCCTGGCACCAGTTTGCGGAGGAGCATCCCGGCGTGCTGCAGTGGACTCTGCAGTACAGGACCGAGGACGACAACCAGATACAGAACGTGACTTTTGATGGTTCAGTGCATCAGTACAGTTTTCAAGCATTACCGGGCACTGAGCTCCGTGTGCTGGGCTGGAAGGTCCTCGGGATACCGGAGAACTTGTCGGATGTGCCGTGGCAACGAGCGGATCTGGACCGCGTACCAGATGGGTAA
Protein
MKKFKNLPVTWAPARTLLPCVVPDGETLVVRGPPLLLDSDDQNRLKPYVWIKDNKEIKQSLTNVTLEVLQKKDGTSEGVYQCGVRHHGLVLGYPIILKFAFIEKYVSVVPENSTVRAGQPFVLTCNISSGPSASITWLKGEEVISENDRYYYIDNKHQLLILNAKEQDAGVYWCKATNPVLNKSRNSSPALVQVEREEGQTGLAFLPLQQDTTIYVSKGSRVVLPCPVVGWPKPEIVWKLTPSIGRTAEQETIDQVLVLPSMQADLEGVFSCSVDGNNDLVKTYNVTITEPVNITLPPASKQVYRASTVRFNCTATGKPAPIITWYKDGKPLVLAGRKVVLTSFEGNRRELLIRSVTSDDAGVYQCFARNALQERGAWARLEVAGAGAEAPRGVRCAAAGPARVRVYWDLMDADVVAYTVQRSVSQGTMYPGVPRTTNEEIITVNEPLTPYKFQVTAYILTKNNKTIASDPSETVTCQGQGVPIKITRVNDERVVVSWHQFAEEHPGVLQWTLQYRTEDDNQIQNVTFDGSVHQYSFQALPGTELRVLGWKVLGIPENLSDVPWQRADLDRVPDG
Summary
Description
May play a role in anteroposterior axis elongation.
Similarity
Belongs to the immunoglobulin superfamily. DCC family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Glycoprotein
Immunoglobulin domain
Membrane
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Protogenin
Uniprot
H9JV68
A0A2W1B6J0
A0A2H1VA08
A0A194QPT3
A0A212EUR8
A0A194Q7Y9
+ More
A0A1B6CPK8 A0A0A9YRL0 A0A146LZX6 A0A154PAX5 J9K5G9 A0A151ICH3 E2ACC0 A0A232FFW8 T1I3G6 A0A151XAF1 A0A026WYR5 A0A158NH66 A0A3L8DR13 A0A195B7T0 A0A0L7RGL8 K7J2J3 A0A195DYD2 E9ILA7 A0A2A3EDT2 A0A195FGB5 A0A224XB00 A0A0N0U2W3 F4WZW8 A0A2H8TTV7 A0A2S2PYT6 A0A2S2R0E9 E2BAR8 A0A0C9RAR6 A0A0C9Q7N4 A0A210PQC4 K1QDZ3 E0VB26 A0A1W4WN68 A0A1W4WNF4 A0A1W4WM82 A0A1I8M4Q9 A0A315W3T1 A0A2I4BMP9 M3ZWX6 A0A087XDF7 V4B3D1 A0A3P9KPA6 A0A1L8H0F6 A0A1L8GT09 A0A1A7ZPX9 H2V9N8 H2LL58 A0A3B3UKM6 A0A3P9KVP9 A0A3B3XK00 A0A3P9IUK2 H0ZFQ2 Q2EY15 W5LKD3 L5K259 H2LVL0 A0A3P9H6W3 D2XNJ9 Q8JI27 A0A341D954 A0A2R8Q867 A0A3B3C995 Q801M2 A0A0R4IIB3 A0A2Y9NVR6 A0A2Y9HIH3 A0A340WH06 A0A2U4CMM3 H0ZFQ6 A0A0R4IP15 A0A384B646 A0A2U3XKT0 U3K682 A0A2Y9T0U4 M3WJ04 A0A2K5ZSF1 A4JYE1 A0A3B4Y0A0 F1LNL1 Q08CP3 G3GRK7 Q2VWP9 A0A3B3QHH1 F6U1B6 A0A2K6T1I7 F6W492 A0A146QEZ4 A0A3B4VEF8 A0A146QVT3
A0A1B6CPK8 A0A0A9YRL0 A0A146LZX6 A0A154PAX5 J9K5G9 A0A151ICH3 E2ACC0 A0A232FFW8 T1I3G6 A0A151XAF1 A0A026WYR5 A0A158NH66 A0A3L8DR13 A0A195B7T0 A0A0L7RGL8 K7J2J3 A0A195DYD2 E9ILA7 A0A2A3EDT2 A0A195FGB5 A0A224XB00 A0A0N0U2W3 F4WZW8 A0A2H8TTV7 A0A2S2PYT6 A0A2S2R0E9 E2BAR8 A0A0C9RAR6 A0A0C9Q7N4 A0A210PQC4 K1QDZ3 E0VB26 A0A1W4WN68 A0A1W4WNF4 A0A1W4WM82 A0A1I8M4Q9 A0A315W3T1 A0A2I4BMP9 M3ZWX6 A0A087XDF7 V4B3D1 A0A3P9KPA6 A0A1L8H0F6 A0A1L8GT09 A0A1A7ZPX9 H2V9N8 H2LL58 A0A3B3UKM6 A0A3P9KVP9 A0A3B3XK00 A0A3P9IUK2 H0ZFQ2 Q2EY15 W5LKD3 L5K259 H2LVL0 A0A3P9H6W3 D2XNJ9 Q8JI27 A0A341D954 A0A2R8Q867 A0A3B3C995 Q801M2 A0A0R4IIB3 A0A2Y9NVR6 A0A2Y9HIH3 A0A340WH06 A0A2U4CMM3 H0ZFQ6 A0A0R4IP15 A0A384B646 A0A2U3XKT0 U3K682 A0A2Y9T0U4 M3WJ04 A0A2K5ZSF1 A4JYE1 A0A3B4Y0A0 F1LNL1 Q08CP3 G3GRK7 Q2VWP9 A0A3B3QHH1 F6U1B6 A0A2K6T1I7 F6W492 A0A146QEZ4 A0A3B4VEF8 A0A146QVT3
Pubmed
19121390
28756777
26354079
22118469
25401762
26823975
+ More
20798317 28648823 24508170 21347285 30249741 20075255 21282665 21719571 28812685 22992520 20566863 25315136 29703783 23542700 23254933 17554307 27762356 21551351 20360741 16881056 16141072 15489334 25329095 23258410 23594743 29451363 15081375 17975172 15057822 21804562 29240929 20431018
20798317 28648823 24508170 21347285 30249741 20075255 21282665 21719571 28812685 22992520 20566863 25315136 29703783 23542700 23254933 17554307 27762356 21551351 20360741 16881056 16141072 15489334 25329095 23258410 23594743 29451363 15081375 17975172 15057822 21804562 29240929 20431018
EMBL
BABH01037400
BABH01037401
KZ150271
PZC71672.1
ODYU01001451
SOQ37667.1
+ More
KQ461194 KPJ06975.1 AGBW02012312 OWR45238.1 KQ459579 KPI99525.1 GEDC01022065 JAS15233.1 GBHO01007967 JAG35637.1 GDHC01006552 JAQ12077.1 KQ434864 KZC08991.1 ABLF02020512 ABLF02020513 ABLF02020514 ABLF02020516 ABLF02046511 KQ978053 KYM97776.1 GL438491 EFN68910.1 NNAY01000289 OXU29453.1 ACPB03023078 KQ982339 KYQ57345.1 KK107063 EZA61185.1 ADTU01015151 QOIP01000005 RLU22847.1 KQ976574 KYM80302.1 KQ414596 KOC69974.1 KQ980107 KYN17624.1 GL764074 EFZ18650.1 KZ288292 PBC29171.1 KQ981625 KYN39059.1 GFTR01006850 JAW09576.1 KQ436008 KOX67608.1 GL888480 EGI60258.1 GFXV01005832 MBW17637.1 GGMS01001360 MBY70563.1 GGMS01014263 MBY83466.1 GL446831 EFN87203.1 GBYB01010177 JAG79944.1 GBYB01010178 JAG79945.1 NEDP02005557 OWF38700.1 JH818408 EKC35117.1 DS235021 EEB10582.1 NHOQ01000384 PWA30475.1 AYCK01018413 KB200347 ESP01846.1 CM004470 OCT89559.1 CM004471 OCT86968.1 HADY01005570 SBP44055.1 ABQF01016568 DQ360114 AY630257 AK039115 AK083540 BC116336 BC116337 AAI16337.1 AAU05740.1 ABC96181.1 BAC30243.1 BAC38947.1 KB031042 ELK05392.1 GU224220 ADB22629.1 AF394058 AAM88671.1 AL935050 BX927325 BX927344 CU463832 AY082380 AAL92552.1 AGTO01020922 AANG04001761 CU458829 CAM60069.1 AABR07070674 AABR07070675 AABR07070676 AABR07070677 AABR07073420 AC141190 BC124155 AAI24156.1 JH000003 EGV99883.1 AY630256 AAU05739.1 AAMC01079933 GCES01132112 JAQ54210.1 GCES01126123 JAQ60199.1
KQ461194 KPJ06975.1 AGBW02012312 OWR45238.1 KQ459579 KPI99525.1 GEDC01022065 JAS15233.1 GBHO01007967 JAG35637.1 GDHC01006552 JAQ12077.1 KQ434864 KZC08991.1 ABLF02020512 ABLF02020513 ABLF02020514 ABLF02020516 ABLF02046511 KQ978053 KYM97776.1 GL438491 EFN68910.1 NNAY01000289 OXU29453.1 ACPB03023078 KQ982339 KYQ57345.1 KK107063 EZA61185.1 ADTU01015151 QOIP01000005 RLU22847.1 KQ976574 KYM80302.1 KQ414596 KOC69974.1 KQ980107 KYN17624.1 GL764074 EFZ18650.1 KZ288292 PBC29171.1 KQ981625 KYN39059.1 GFTR01006850 JAW09576.1 KQ436008 KOX67608.1 GL888480 EGI60258.1 GFXV01005832 MBW17637.1 GGMS01001360 MBY70563.1 GGMS01014263 MBY83466.1 GL446831 EFN87203.1 GBYB01010177 JAG79944.1 GBYB01010178 JAG79945.1 NEDP02005557 OWF38700.1 JH818408 EKC35117.1 DS235021 EEB10582.1 NHOQ01000384 PWA30475.1 AYCK01018413 KB200347 ESP01846.1 CM004470 OCT89559.1 CM004471 OCT86968.1 HADY01005570 SBP44055.1 ABQF01016568 DQ360114 AY630257 AK039115 AK083540 BC116336 BC116337 AAI16337.1 AAU05740.1 ABC96181.1 BAC30243.1 BAC38947.1 KB031042 ELK05392.1 GU224220 ADB22629.1 AF394058 AAM88671.1 AL935050 BX927325 BX927344 CU463832 AY082380 AAL92552.1 AGTO01020922 AANG04001761 CU458829 CAM60069.1 AABR07070674 AABR07070675 AABR07070676 AABR07070677 AABR07073420 AC141190 BC124155 AAI24156.1 JH000003 EGV99883.1 AY630256 AAU05739.1 AAMC01079933 GCES01132112 JAQ54210.1 GCES01126123 JAQ60199.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000076502
UP000007819
+ More
UP000078542 UP000000311 UP000215335 UP000015103 UP000075809 UP000053097 UP000005205 UP000279307 UP000078540 UP000053825 UP000002358 UP000078492 UP000242457 UP000078541 UP000053105 UP000007755 UP000008237 UP000242188 UP000005408 UP000009046 UP000192223 UP000095301 UP000192220 UP000002852 UP000028760 UP000030746 UP000265180 UP000186698 UP000005226 UP000001038 UP000261500 UP000261480 UP000265200 UP000007754 UP000000589 UP000018467 UP000010552 UP000252040 UP000000437 UP000261560 UP000248483 UP000248481 UP000265300 UP000245320 UP000245341 UP000016665 UP000248484 UP000011712 UP000233140 UP000261360 UP000002494 UP000001075 UP000261540 UP000008143 UP000233220 UP000008225 UP000261420
UP000078542 UP000000311 UP000215335 UP000015103 UP000075809 UP000053097 UP000005205 UP000279307 UP000078540 UP000053825 UP000002358 UP000078492 UP000242457 UP000078541 UP000053105 UP000007755 UP000008237 UP000242188 UP000005408 UP000009046 UP000192223 UP000095301 UP000192220 UP000002852 UP000028760 UP000030746 UP000265180 UP000186698 UP000005226 UP000001038 UP000261500 UP000261480 UP000265200 UP000007754 UP000000589 UP000018467 UP000010552 UP000252040 UP000000437 UP000261560 UP000248483 UP000248481 UP000265300 UP000245320 UP000245341 UP000016665 UP000248484 UP000011712 UP000233140 UP000261360 UP000002494 UP000001075 UP000261540 UP000008143 UP000233220 UP000008225 UP000261420
Pfam
Interpro
IPR036116
FN3_sf
+ More
IPR003961 FN3_dom
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR013106 Ig_V-set
IPR013098 Ig_I-set
IPR013151 Immunoglobulin
IPR029000 Cyclophilin-like_dom_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR033011 Protogenin
IPR032983 CDO
IPR010560 Neogenin_C
IPR033024 Neogenin
IPR003961 FN3_dom
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR013106 Ig_V-set
IPR013098 Ig_I-set
IPR013151 Immunoglobulin
IPR029000 Cyclophilin-like_dom_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR033011 Protogenin
IPR032983 CDO
IPR010560 Neogenin_C
IPR033024 Neogenin
Gene 3D
CDD
ProteinModelPortal
H9JV68
A0A2W1B6J0
A0A2H1VA08
A0A194QPT3
A0A212EUR8
A0A194Q7Y9
+ More
A0A1B6CPK8 A0A0A9YRL0 A0A146LZX6 A0A154PAX5 J9K5G9 A0A151ICH3 E2ACC0 A0A232FFW8 T1I3G6 A0A151XAF1 A0A026WYR5 A0A158NH66 A0A3L8DR13 A0A195B7T0 A0A0L7RGL8 K7J2J3 A0A195DYD2 E9ILA7 A0A2A3EDT2 A0A195FGB5 A0A224XB00 A0A0N0U2W3 F4WZW8 A0A2H8TTV7 A0A2S2PYT6 A0A2S2R0E9 E2BAR8 A0A0C9RAR6 A0A0C9Q7N4 A0A210PQC4 K1QDZ3 E0VB26 A0A1W4WN68 A0A1W4WNF4 A0A1W4WM82 A0A1I8M4Q9 A0A315W3T1 A0A2I4BMP9 M3ZWX6 A0A087XDF7 V4B3D1 A0A3P9KPA6 A0A1L8H0F6 A0A1L8GT09 A0A1A7ZPX9 H2V9N8 H2LL58 A0A3B3UKM6 A0A3P9KVP9 A0A3B3XK00 A0A3P9IUK2 H0ZFQ2 Q2EY15 W5LKD3 L5K259 H2LVL0 A0A3P9H6W3 D2XNJ9 Q8JI27 A0A341D954 A0A2R8Q867 A0A3B3C995 Q801M2 A0A0R4IIB3 A0A2Y9NVR6 A0A2Y9HIH3 A0A340WH06 A0A2U4CMM3 H0ZFQ6 A0A0R4IP15 A0A384B646 A0A2U3XKT0 U3K682 A0A2Y9T0U4 M3WJ04 A0A2K5ZSF1 A4JYE1 A0A3B4Y0A0 F1LNL1 Q08CP3 G3GRK7 Q2VWP9 A0A3B3QHH1 F6U1B6 A0A2K6T1I7 F6W492 A0A146QEZ4 A0A3B4VEF8 A0A146QVT3
A0A1B6CPK8 A0A0A9YRL0 A0A146LZX6 A0A154PAX5 J9K5G9 A0A151ICH3 E2ACC0 A0A232FFW8 T1I3G6 A0A151XAF1 A0A026WYR5 A0A158NH66 A0A3L8DR13 A0A195B7T0 A0A0L7RGL8 K7J2J3 A0A195DYD2 E9ILA7 A0A2A3EDT2 A0A195FGB5 A0A224XB00 A0A0N0U2W3 F4WZW8 A0A2H8TTV7 A0A2S2PYT6 A0A2S2R0E9 E2BAR8 A0A0C9RAR6 A0A0C9Q7N4 A0A210PQC4 K1QDZ3 E0VB26 A0A1W4WN68 A0A1W4WNF4 A0A1W4WM82 A0A1I8M4Q9 A0A315W3T1 A0A2I4BMP9 M3ZWX6 A0A087XDF7 V4B3D1 A0A3P9KPA6 A0A1L8H0F6 A0A1L8GT09 A0A1A7ZPX9 H2V9N8 H2LL58 A0A3B3UKM6 A0A3P9KVP9 A0A3B3XK00 A0A3P9IUK2 H0ZFQ2 Q2EY15 W5LKD3 L5K259 H2LVL0 A0A3P9H6W3 D2XNJ9 Q8JI27 A0A341D954 A0A2R8Q867 A0A3B3C995 Q801M2 A0A0R4IIB3 A0A2Y9NVR6 A0A2Y9HIH3 A0A340WH06 A0A2U4CMM3 H0ZFQ6 A0A0R4IP15 A0A384B646 A0A2U3XKT0 U3K682 A0A2Y9T0U4 M3WJ04 A0A2K5ZSF1 A4JYE1 A0A3B4Y0A0 F1LNL1 Q08CP3 G3GRK7 Q2VWP9 A0A3B3QHH1 F6U1B6 A0A2K6T1I7 F6W492 A0A146QEZ4 A0A3B4VEF8 A0A146QVT3
PDB
5E4I
E-value=2.1476e-23,
Score=271
Ontologies
GO
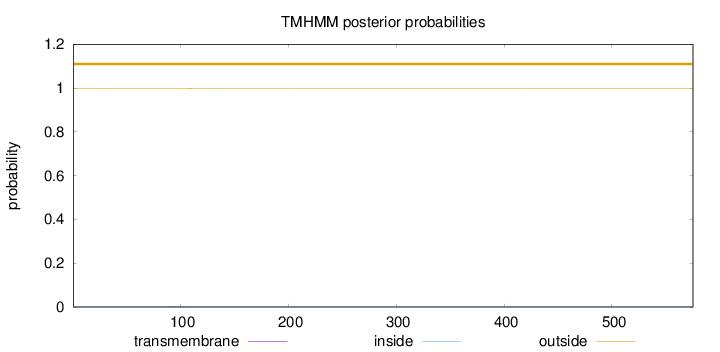
Topology
Subcellular location
Membrane
Length:
575
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0516500000000001
Exp number, first 60 AAs:
0.00293
Total prob of N-in:
0.00370
outside
1 - 575
Population Genetic Test Statistics
Pi
213.134241
Theta
167.477516
Tajima's D
0.369528
CLR
0.326741
CSRT
0.477526123693815
Interpretation
Uncertain
