Gene
KWMTBOMO03145
Pre Gene Modal
BGIBMGA013475
Annotation
PREDICTED:_mothers_against_decapentaplegic_homolog_6_[Amyelois_transitella]
Full name
Mothers against decapentaplegic homolog
+ More
Mothers against decapentaplegic homolog 6
Mothers against decapentaplegic homolog 6
Alternative Name
SMAD family member
SMAD family member 6
Mad homolog 7
SMAD family member 6
Mad homolog 7
Transcription factor
Location in the cell
Mitochondrial Reliability : 1.123 Nuclear Reliability : 1.618
Sequence
CDS
ATGTTCCGGCGTCGCGCGCTGCGTCGGCGGCTGGCGTCGCTAGGAGTAGCGGCGGTGCCTGACGACGTGCTGCGAAGGCTGGCGCGGGCGCTGGACGCGGGGCCCGGCGGTGCCCTGTGCGTACCGCCCGCGCCGCCTGATGACCGTAGGCTGCGCCTCGCGCTCGCCAGGGCCTTCTGCTTCCCAGACCTGCGCGACCTGTCCGAGCTGCGTCGGATGCCTCACTGCACCGACAATTCCTGCTGCAACCCTTACCACTGGAGTCGTCTCTGCAAACCCGAAACCCCCCCGCCGCCATACACTCGTTTTGCCATGGATGACTATAAACCTAAAGATCACGGAGAGAGCACGGAAGACGCGCGGAGGACAGACCACGAGCGGTTTGACTCTGGCTCCATGGCTACTGACGGAGAAGAGCGTCAGAGTTGGGAGACAGAGTGGTGTCGGCTCGCATACTGGGAGCTGACGCAGCGCGTGGGCCGTCAGTTCGGCGTGAGGATGCGCGCCGTGGACGTGTTCGGCGGAGGAGGCGGCGCGTGCGGCTCCGGCCGGCACGGCCTGTGTCTGGACGCGCTCGACCGCGTCGACCACCGCCCCGACCTGGCGCAGCCCGACCAGGTCCGCAAGACTCGTGCCAAGATAGGGCTGGGCGTGACGCTGTCGCTGGAGGGCGACGGCGTGTGGGTGTACAACCGCGGCGCCGAGCCGGTGTTCGTGTCGTCGCCGGCGCTGGACGCGGCGGCGGCGCGGGCGCTGCTGGTGTGGCGCGTGGCGGCCGGCCACTGTCTGCGCGTGTTCCCGCCGCCCCCCTTCGCCGCGCCGCGCCCCGCCCCGCCGCGCCCCGCCCTGCCGCGCGACCCCCGCTCGGTGCGCATCTCGTTCGCGAAGGGCTGGGGGCCAAAGTACTCGCGACGTGACGTCACAGCGTGCCCGTGTTGGCTTGAGGTGCTGCTGGCGCCGCCCAGCTGA
Protein
MFRRRALRRRLASLGVAAVPDDVLRRLARALDAGPGGALCVPPAPPDDRRLRLALARAFCFPDLRDLSELRRMPHCTDNSCCNPYHWSRLCKPETPPPPYTRFAMDDYKPKDHGESTEDARRTDHERFDSGSMATDGEERQSWETEWCRLAYWELTQRVGRQFGVRMRAVDVFGGGGGACGSGRHGLCLDALDRVDHRPDLAQPDQVRKTRAKIGLGVTLSLEGDGVWVYNRGAEPVFVSSPALDAAAARALLVWRVAAGHCLRVFPPPPFAAPRPAPPRPALPRDPRSVRISFAKGWGPKYSRRDVTACPCWLEVLLAPPS
Summary
Description
Acts as a mediator of TGF-beta and BMP antiflammatory activity. Suppresses IL1R-TLR signaling through its direct interaction with PEL1, preventing NF-kappa-B activation, nuclear transport and NF-kappa-B-mediated expression of proinflammatory genes. May block the BMP-SMAD1 signaling pathway by competing with SMAD4 for receptor-activated SMAD1-binding. Binds to regulatory elements in target promoter regions.
Binds to regulatory elements in target promoter regions (By similarity). May block the BMP-SMAD1 signaling pathway by competing with SMAD4 for receptor-activated SMAD1-binding (By similarity). Acts as a mediator of TGF-beta and BMP antiflammatory activity. Suppresses IL1R-TLR signaling through its direct interaction with PEL1, preventing NF-kappa-B activation, nuclear transport and NF-kappa-B-mediated expression of proinflammatory genes.
Acts as a mediator of TGF-beta and BMP antiflammatory activity. Suppresses IL1R-TLR signaling, preventing NF-kappa-B activation, and BMP-SMAD1 signaling pathway. Binds to regulatory elements in target promoter regions (By similarity).
Binds to regulatory elements in target promoter regions (By similarity). May block the BMP-SMAD1 signaling pathway by competing with SMAD4 for receptor-activated SMAD1-binding (By similarity). Acts as a mediator of TGF-beta and BMP antiflammatory activity. Suppresses IL1R-TLR signaling through its direct interaction with PEL1, preventing NF-kappa-B activation, nuclear transport and NF-kappa-B-mediated expression of proinflammatory genes.
Acts as a mediator of TGF-beta and BMP antiflammatory activity. Suppresses IL1R-TLR signaling, preventing NF-kappa-B activation, and BMP-SMAD1 signaling pathway. Binds to regulatory elements in target promoter regions (By similarity).
Subunit
Interacts with NEDD4L (By similarity). Interacts with WWP1 (By similarity). Interacts with STAMBP and PRKX. Interacts with RNF111 and AXIN1. Interacts with TGF-beta type I receptor superfamily members, including ACVR1B, BMPR1B and TGFBR1. In response to BMP2, but not to TGFB treatment, interacts with SMAD1, but not with SMAD2, nor with SMAD4; this interaction may inhibit SMAD1 binding to SMAD4. Interacts with HOXC8 and HOXC9. Interacts with PELI1; this interaction interferes with PELI1 complex formation with TRAF6, IRAK1, IRAK4 and MYD88 in response to IL1B and hence negatively regulates IL1R-TLR signaling.
Interacts with NEDD4L. Interacts with WWP1. Interacts with STAMBP and PRKX (By similarity). Interacts with RNF111 and AXIN1. Interacts with TGF-beta type I receptor superfamily members, including ACVR1B, BMPR1B and TGFBR1. In response to BMP2 treatment, interacts with SMAD1; this interaction may inhibit SMAD1-binding to SMAD4. Interacts with HOXC8 and HOXC9 (By similarity). Interacts with PELI1; this interaction interferes with PELI1 complex formation with TRAF6, IRAK1, IRAK4 and MYD88 in response to IL1B and hence negatively regulates IL1R-TLR signaling.
Interacts with NEDD4L. Interacts with WWP1. Interacts with STAMBP and PRKX (By similarity). Interacts with RNF111 and AXIN1. Interacts with TGF-beta type I receptor superfamily members, including ACVR1B, BMPR1B and TGFBR1. In response to BMP2 treatment, interacts with SMAD1; this interaction may inhibit SMAD1-binding to SMAD4. Interacts with HOXC8 and HOXC9 (By similarity). Interacts with PELI1; this interaction interferes with PELI1 complex formation with TRAF6, IRAK1, IRAK4 and MYD88 in response to IL1B and hence negatively regulates IL1R-TLR signaling.
Similarity
Belongs to the dwarfin/SMAD family.
Keywords
Alternative splicing
Complete proteome
Craniosynostosis
Disease mutation
DNA-binding
Isopeptide bond
Metal-binding
Methylation
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Transcription
Transcription regulation
Ubl conjugation
Zinc
Feature
chain Mothers against decapentaplegic homolog
splice variant In isoform B.
sequence variant In CRS7; associated with disease susceptibility.
splice variant In isoform B.
sequence variant In CRS7; associated with disease susceptibility.
Uniprot
A0A2A4JL28
A0A194Q1S7
A0A067QH14
A0A1B6D1W7
A0A1B6KBR2
A0A1B6HX09
+ More
A0A1B6FLX6 A0A2H8TWX8 J9KAL4 A0A1W4WBD3 A0A088AG08 A0A158NLE2 A0A1Y1KHV7 A0A2S2R9Z4 A0A0L7QLV9 A0A2S2QEW2 A0A195CZP9 A0A3L8D728 A0A154NYQ8 A0A195F1C0 A0A151XCF7 A0A0A9WFP9 A0A0K8S971 A0A1S3EPF8 F6ZM58 A0A383YTH8 H2NNK1 G9KQ25 A0A1D5QSL6 F6RB41 A0A2K5VS10 A0A1D5Q8K4 G3RB41 G3W192 I3LX61 A0A2K6M3E1 A0A218V124 A0A2I3HZI2 U6DB10 U3KEF4 A0A2K5KI94 A0A1S3K878 A0A2J8UC48 A0A2J8NR46 O43541 A0A2K6CDL1 A0A0D9RLC6 A0A1A8RIW1 A0A2K6EJ18 K6ZBV0 A0A2K5QQR8 A0A2K5C913 M3WGP4 A0A2I0MD85 U3B961 A0A2R8MQJ0 A0A2Y9HJN0 K7CVP9 A0A151M8V2 A0A1U8C050 E1BH37 A0A2Y9FP97 O35182 A0A340WME7 F1SJJ2 A0A2Y9NWK6 L5K1U7 L5LF53 A0A2U3WYJ5 H0XD56 Q80ZV9 A0A2I3NFA6 A0A2Y9K338 M3XMK5 F6RFA7 W5QA49 A0A1D5PB14 A0A3L8SK14 Q9W734 C3U1W5 A0A0Q3UPN4 A0A2Y9FZG9 A0A1V4JE69 G3TB12 G3T436 A0A1A8K1D1 A0A1A8PC92 A0A1A8I2H9 A0A1A8PBR8 A0A1A8D859 A0A1A7ZSU1 H3A8S2 A0A0S7HG99 A0A3P8TU61
A0A1B6FLX6 A0A2H8TWX8 J9KAL4 A0A1W4WBD3 A0A088AG08 A0A158NLE2 A0A1Y1KHV7 A0A2S2R9Z4 A0A0L7QLV9 A0A2S2QEW2 A0A195CZP9 A0A3L8D728 A0A154NYQ8 A0A195F1C0 A0A151XCF7 A0A0A9WFP9 A0A0K8S971 A0A1S3EPF8 F6ZM58 A0A383YTH8 H2NNK1 G9KQ25 A0A1D5QSL6 F6RB41 A0A2K5VS10 A0A1D5Q8K4 G3RB41 G3W192 I3LX61 A0A2K6M3E1 A0A218V124 A0A2I3HZI2 U6DB10 U3KEF4 A0A2K5KI94 A0A1S3K878 A0A2J8UC48 A0A2J8NR46 O43541 A0A2K6CDL1 A0A0D9RLC6 A0A1A8RIW1 A0A2K6EJ18 K6ZBV0 A0A2K5QQR8 A0A2K5C913 M3WGP4 A0A2I0MD85 U3B961 A0A2R8MQJ0 A0A2Y9HJN0 K7CVP9 A0A151M8V2 A0A1U8C050 E1BH37 A0A2Y9FP97 O35182 A0A340WME7 F1SJJ2 A0A2Y9NWK6 L5K1U7 L5LF53 A0A2U3WYJ5 H0XD56 Q80ZV9 A0A2I3NFA6 A0A2Y9K338 M3XMK5 F6RFA7 W5QA49 A0A1D5PB14 A0A3L8SK14 Q9W734 C3U1W5 A0A0Q3UPN4 A0A2Y9FZG9 A0A1V4JE69 G3TB12 G3T436 A0A1A8K1D1 A0A1A8PC92 A0A1A8I2H9 A0A1A8PBR8 A0A1A8D859 A0A1A7ZSU1 H3A8S2 A0A0S7HG99 A0A3P8TU61
Pubmed
26354079
24845553
21347285
28004739
30249741
25401762
+ More
26823975 19892987 23236062 17431167 22398555 21709235 8673135 9436979 9712726 15489334 9759503 10647776 10708948 10708949 10722652 11284962 11483516 14657019 16601693 16951688 16491121 22275001 23455153 27606499 16136131 17975172 23371554 25243066 22293439 19393038 9335505 16141072 15221015 15496141 23747011 23610558 30723633 23258410 17495919 20809919 15592404 30282656 10525349 19352514 9215903
26823975 19892987 23236062 17431167 22398555 21709235 8673135 9436979 9712726 15489334 9759503 10647776 10708948 10708949 10722652 11284962 11483516 14657019 16601693 16951688 16491121 22275001 23455153 27606499 16136131 17975172 23371554 25243066 22293439 19393038 9335505 16141072 15221015 15496141 23747011 23610558 30723633 23258410 17495919 20809919 15592404 30282656 10525349 19352514 9215903
EMBL
NWSH01001073
PCG72757.1
KQ459579
KPI99532.1
KK853452
KDR07534.1
+ More
GEDC01017990 GEDC01017599 GEDC01000058 JAS19308.1 JAS19699.1 JAS37240.1 GEBQ01031122 JAT08855.1 GECU01028485 JAS79221.1 GECZ01018572 JAS51197.1 GFXV01006952 MBW18757.1 ABLF02021478 ADTU01019326 ADTU01019327 ADTU01019328 ADTU01019329 ADTU01019330 ADTU01019331 ADTU01019332 ADTU01019333 GEZM01086045 JAV59820.1 GGMS01016969 MBY86172.1 KQ414914 KOC59534.1 GGMS01007063 MBY76266.1 KQ977115 KYN05614.1 QOIP01000012 RLU16074.1 KQ434773 KZC04010.1 KQ981864 KYN34370.1 KQ982314 KYQ58027.1 GBHO01037378 GBHO01031713 GDHC01002996 JAG06226.1 JAG11891.1 JAQ15633.1 GBRD01015999 JAG49827.1 ABGA01218410 ABGA01218411 ABGA01218412 ABGA01218413 AC188093 JP018406 AES07004.1 JSUE03038580 AQIA01063082 CABD030096845 CABD030096846 AEFK01007919 AEFK01007920 AEFK01007921 AEFK01007922 AGTP01003289 AGTP01003290 MUZQ01000079 OWK59420.1 ADFV01035931 ADFV01035932 ADFV01035933 ADFV01035934 ADFV01035935 ADFV01035936 HAAF01007987 CCP79811.1 AGTO01020820 NDHI03003463 PNJ42857.1 NBAG03000224 PNI74239.1 U59914 AF035528 AF043640 AF037469 AF041065 AF041062 AF041063 AF041064 AM909653 BC012986 AF101474 AQIB01139235 AQIB01139236 HAEI01008661 SBS05632.1 AACZ04064628 AACZ04064629 GABC01009282 GABF01008990 GABE01005911 JAA02056.1 JAA13155.1 JAA38828.1 AANG04001761 AKCR02000019 PKK27639.1 GAMT01009747 JAB02114.1 GABD01004337 GABD01004335 JAA28765.1 AKHW03006295 KYO20946.1 AF010133 AK004671 AEMK02000004 DQIR01023642 HCZ79117.1 KB031042 ELK05307.1 KB112376 ELK24837.1 AAQR03026100 AAQR03026101 AAQR03026102 AAQR03026103 BC047280 AAH47280.1 AHZZ02031197 AEYP01046988 AMGL01101877 AMGL01101878 AADN05000047 QUSF01000015 RLW03532.1 AF165889 FJ417094 ACO82015.1 LMAW01003223 KQK73306.1 LSYS01007908 OPJ70325.1 HAEE01006016 SBR26036.1 HAEF01013011 HAEG01007335 SBR79005.1 HAED01005744 SBQ91774.1 HAEH01006250 SBR78721.1 HADZ01020883 HAEA01001827 SBQ30307.1 HADY01006550 HAEJ01003687 SBP45035.1 AFYH01123852 AFYH01123853 AFYH01123854 GBYX01441391 JAO39983.1
GEDC01017990 GEDC01017599 GEDC01000058 JAS19308.1 JAS19699.1 JAS37240.1 GEBQ01031122 JAT08855.1 GECU01028485 JAS79221.1 GECZ01018572 JAS51197.1 GFXV01006952 MBW18757.1 ABLF02021478 ADTU01019326 ADTU01019327 ADTU01019328 ADTU01019329 ADTU01019330 ADTU01019331 ADTU01019332 ADTU01019333 GEZM01086045 JAV59820.1 GGMS01016969 MBY86172.1 KQ414914 KOC59534.1 GGMS01007063 MBY76266.1 KQ977115 KYN05614.1 QOIP01000012 RLU16074.1 KQ434773 KZC04010.1 KQ981864 KYN34370.1 KQ982314 KYQ58027.1 GBHO01037378 GBHO01031713 GDHC01002996 JAG06226.1 JAG11891.1 JAQ15633.1 GBRD01015999 JAG49827.1 ABGA01218410 ABGA01218411 ABGA01218412 ABGA01218413 AC188093 JP018406 AES07004.1 JSUE03038580 AQIA01063082 CABD030096845 CABD030096846 AEFK01007919 AEFK01007920 AEFK01007921 AEFK01007922 AGTP01003289 AGTP01003290 MUZQ01000079 OWK59420.1 ADFV01035931 ADFV01035932 ADFV01035933 ADFV01035934 ADFV01035935 ADFV01035936 HAAF01007987 CCP79811.1 AGTO01020820 NDHI03003463 PNJ42857.1 NBAG03000224 PNI74239.1 U59914 AF035528 AF043640 AF037469 AF041065 AF041062 AF041063 AF041064 AM909653 BC012986 AF101474 AQIB01139235 AQIB01139236 HAEI01008661 SBS05632.1 AACZ04064628 AACZ04064629 GABC01009282 GABF01008990 GABE01005911 JAA02056.1 JAA13155.1 JAA38828.1 AANG04001761 AKCR02000019 PKK27639.1 GAMT01009747 JAB02114.1 GABD01004337 GABD01004335 JAA28765.1 AKHW03006295 KYO20946.1 AF010133 AK004671 AEMK02000004 DQIR01023642 HCZ79117.1 KB031042 ELK05307.1 KB112376 ELK24837.1 AAQR03026100 AAQR03026101 AAQR03026102 AAQR03026103 BC047280 AAH47280.1 AHZZ02031197 AEYP01046988 AMGL01101877 AMGL01101878 AADN05000047 QUSF01000015 RLW03532.1 AF165889 FJ417094 ACO82015.1 LMAW01003223 KQK73306.1 LSYS01007908 OPJ70325.1 HAEE01006016 SBR26036.1 HAEF01013011 HAEG01007335 SBR79005.1 HAED01005744 SBQ91774.1 HAEH01006250 SBR78721.1 HADZ01020883 HAEA01001827 SBQ30307.1 HADY01006550 HAEJ01003687 SBP45035.1 AFYH01123852 AFYH01123853 AFYH01123854 GBYX01441391 JAO39983.1
Proteomes
UP000218220
UP000053268
UP000027135
UP000007819
UP000192223
UP000005203
+ More
UP000005205 UP000053825 UP000078542 UP000279307 UP000076502 UP000078541 UP000075809 UP000081671 UP000002281 UP000261681 UP000001595 UP000006718 UP000233100 UP000001519 UP000007648 UP000005215 UP000233180 UP000197619 UP000001073 UP000016665 UP000233060 UP000085678 UP000005640 UP000233120 UP000029965 UP000233160 UP000002277 UP000233040 UP000233020 UP000011712 UP000053872 UP000008225 UP000248481 UP000050525 UP000189706 UP000009136 UP000248484 UP000000589 UP000265300 UP000008227 UP000248483 UP000010552 UP000245340 UP000005225 UP000028761 UP000248482 UP000000715 UP000002280 UP000002356 UP000000539 UP000276834 UP000051836 UP000248480 UP000190648 UP000007646 UP000008672 UP000265080
UP000005205 UP000053825 UP000078542 UP000279307 UP000076502 UP000078541 UP000075809 UP000081671 UP000002281 UP000261681 UP000001595 UP000006718 UP000233100 UP000001519 UP000007648 UP000005215 UP000233180 UP000197619 UP000001073 UP000016665 UP000233060 UP000085678 UP000005640 UP000233120 UP000029965 UP000233160 UP000002277 UP000233040 UP000233020 UP000011712 UP000053872 UP000008225 UP000248481 UP000050525 UP000189706 UP000009136 UP000248484 UP000000589 UP000265300 UP000008227 UP000248483 UP000010552 UP000245340 UP000005225 UP000028761 UP000248482 UP000000715 UP000002280 UP000002356 UP000000539 UP000276834 UP000051836 UP000248480 UP000190648 UP000007646 UP000008672 UP000265080
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JL28
A0A194Q1S7
A0A067QH14
A0A1B6D1W7
A0A1B6KBR2
A0A1B6HX09
+ More
A0A1B6FLX6 A0A2H8TWX8 J9KAL4 A0A1W4WBD3 A0A088AG08 A0A158NLE2 A0A1Y1KHV7 A0A2S2R9Z4 A0A0L7QLV9 A0A2S2QEW2 A0A195CZP9 A0A3L8D728 A0A154NYQ8 A0A195F1C0 A0A151XCF7 A0A0A9WFP9 A0A0K8S971 A0A1S3EPF8 F6ZM58 A0A383YTH8 H2NNK1 G9KQ25 A0A1D5QSL6 F6RB41 A0A2K5VS10 A0A1D5Q8K4 G3RB41 G3W192 I3LX61 A0A2K6M3E1 A0A218V124 A0A2I3HZI2 U6DB10 U3KEF4 A0A2K5KI94 A0A1S3K878 A0A2J8UC48 A0A2J8NR46 O43541 A0A2K6CDL1 A0A0D9RLC6 A0A1A8RIW1 A0A2K6EJ18 K6ZBV0 A0A2K5QQR8 A0A2K5C913 M3WGP4 A0A2I0MD85 U3B961 A0A2R8MQJ0 A0A2Y9HJN0 K7CVP9 A0A151M8V2 A0A1U8C050 E1BH37 A0A2Y9FP97 O35182 A0A340WME7 F1SJJ2 A0A2Y9NWK6 L5K1U7 L5LF53 A0A2U3WYJ5 H0XD56 Q80ZV9 A0A2I3NFA6 A0A2Y9K338 M3XMK5 F6RFA7 W5QA49 A0A1D5PB14 A0A3L8SK14 Q9W734 C3U1W5 A0A0Q3UPN4 A0A2Y9FZG9 A0A1V4JE69 G3TB12 G3T436 A0A1A8K1D1 A0A1A8PC92 A0A1A8I2H9 A0A1A8PBR8 A0A1A8D859 A0A1A7ZSU1 H3A8S2 A0A0S7HG99 A0A3P8TU61
A0A1B6FLX6 A0A2H8TWX8 J9KAL4 A0A1W4WBD3 A0A088AG08 A0A158NLE2 A0A1Y1KHV7 A0A2S2R9Z4 A0A0L7QLV9 A0A2S2QEW2 A0A195CZP9 A0A3L8D728 A0A154NYQ8 A0A195F1C0 A0A151XCF7 A0A0A9WFP9 A0A0K8S971 A0A1S3EPF8 F6ZM58 A0A383YTH8 H2NNK1 G9KQ25 A0A1D5QSL6 F6RB41 A0A2K5VS10 A0A1D5Q8K4 G3RB41 G3W192 I3LX61 A0A2K6M3E1 A0A218V124 A0A2I3HZI2 U6DB10 U3KEF4 A0A2K5KI94 A0A1S3K878 A0A2J8UC48 A0A2J8NR46 O43541 A0A2K6CDL1 A0A0D9RLC6 A0A1A8RIW1 A0A2K6EJ18 K6ZBV0 A0A2K5QQR8 A0A2K5C913 M3WGP4 A0A2I0MD85 U3B961 A0A2R8MQJ0 A0A2Y9HJN0 K7CVP9 A0A151M8V2 A0A1U8C050 E1BH37 A0A2Y9FP97 O35182 A0A340WME7 F1SJJ2 A0A2Y9NWK6 L5K1U7 L5LF53 A0A2U3WYJ5 H0XD56 Q80ZV9 A0A2I3NFA6 A0A2Y9K338 M3XMK5 F6RFA7 W5QA49 A0A1D5PB14 A0A3L8SK14 Q9W734 C3U1W5 A0A0Q3UPN4 A0A2Y9FZG9 A0A1V4JE69 G3TB12 G3T436 A0A1A8K1D1 A0A1A8PC92 A0A1A8I2H9 A0A1A8PBR8 A0A1A8D859 A0A1A7ZSU1 H3A8S2 A0A0S7HG99 A0A3P8TU61
PDB
1DEV
E-value=2.03661e-09,
Score=148
Ontologies
GO
GO:0003677
GO:0005634
GO:0007179
GO:0005737
GO:0003700
GO:0005667
GO:0006355
GO:0042802
GO:0008285
GO:0070411
GO:0035904
GO:0001657
GO:0003180
GO:0030509
GO:0003682
GO:0003183
GO:0030512
GO:0007352
GO:0031589
GO:0003148
GO:0010991
GO:0003184
GO:1902895
GO:0045444
GO:0005794
GO:0034616
GO:0045668
GO:0031625
GO:0060976
GO:0030617
GO:0016604
GO:0030514
GO:0070698
GO:0000978
GO:0005829
GO:0003281
GO:0043066
GO:0070410
GO:0060394
GO:0034713
GO:0006955
GO:0032496
GO:0070412
GO:0030279
GO:0032991
GO:0000981
GO:0044212
GO:0043627
GO:0046872
GO:0003170
GO:0097756
GO:0045907
GO:0060977
GO:0060948
GO:1990423
GO:0007093
GO:0032331
GO:0010693
GO:1902832
GO:0050767
GO:1902844
GO:0005768
GO:0005622
GO:0005515
GO:0016021
GO:0045211
GO:0006281
GO:0048384
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
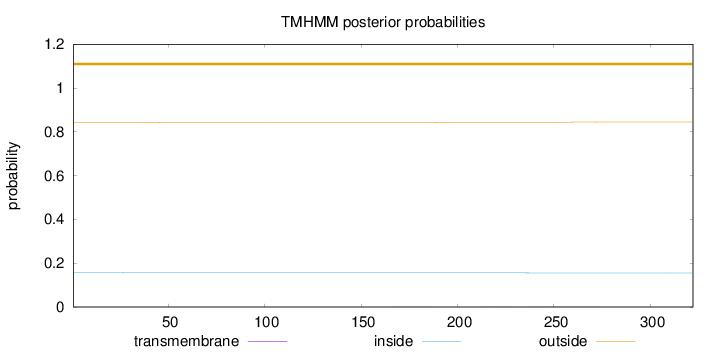
Length:
322
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03839
Exp number, first 60 AAs:
0.0036
Total prob of N-in:
0.15693
outside
1 - 322
Population Genetic Test Statistics
Pi
208.862164
Theta
187.210304
Tajima's D
0.502859
CLR
0.749169
CSRT
0.51982400879956
Interpretation
Uncertain
