Gene
KWMTBOMO03143
Pre Gene Modal
BGIBMGA013477
Annotation
PREDICTED:_synaptic_vesicle_glycoprotein_2B-like_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 4.918
Sequence
CDS
ATGGAGGAGCACGGAGAGAGCAAAGATCGAGTGCAGAGGGCGCCCTTCGAGACCGCGCTGCATCACGCCGGTTACGGATGCTTCCAGTGGCTGCTGCTGCTGTCCTGCGGCGCCGTGTACGCCGTCTGCGCACTCAGCACGACCACGCTCAGCTTCGTGCTGCCCGCCACCAAGAATGACTTCGAGCTGTCGTCCTCCGACAAGGGCCGCCTCACCGCCACGCCGCTCATCGGAATGTGCGTCGGCTCGTACTTCTGGGGTAACTTGGCGGACGCTCGAGGCCGAAAGATCGCCATCATTGGAGCTCTACTTTTGGATGCTCTGGCCGCGTTCCTGTCCAGCGTTGTGCAGTCCTTCTCCGGGTTCCTGGCTTGCAGGTTCTTCTCCGGGTTCGGAATCATCGGAGCCACGGGCCTCATCTTCCCTTACTTCGGTGACTTTCTGTCGTTGAGGCACAGAGACGTGATGCTGTGCAGGCTCGAGGTGTTTTGGACCGTGGGAACTATACTATTGCCAGGGATCGCGTTTCTGATCATCCCGGACAAGCGGTTCACACTGACGGCGGCCGACGCTGACTTCCAGTACACGTCGTGGCGCGTGTTCGTGGCGGCGTGCGGCGTGCCCAGCCTGCTCGTGGTGCTGCTTCTGCTGCCGTTCCCGGAGAGCCCGCGCTACCTGCTGCACGCAAATCGCGCCGAGCAGGCGCTCCACGTGCTGCGCAGGATCTTTGTCGTCAATACCAAGCTTGACGCCAAAGACTTCCCGGTCGAATCGATGATTAGTGCGGGGGACGGCGATGAGGAGGAGGTGGTGTCTGCCGACAACAACGGCAACGAGACCAAGGCACCGCTCACTTGGAGACAGCGCTGGGACGCTTTCTGGATCCGGAAGAAGATGCTCGTCACGCCCCCGTACATCGGACTGTTAGTCCTGTGCTGTTTCGTGGACTTCGGCCTCATGTTCAGTTACTACACTTTAATGATGTGGTTCCCGGACCTTTTCGAGCGGTTCAGCCAGTTCGCGTCGCTGTACGGAGACGAGCCCGCCGGAGTCTGCGAGGTTTCGCTGGCCGTCGCCAATGCCACGACCGACATACCAATCCAGAGCACGATGTCGCCGGTGCACGACCAGGTGTACATCCACACGCTGGTGGTGGCGCTGAGCTGCGTCCCCACCGCGCTTTGGGTCGGCCTCACAATCAACATCGTCGGCAAGAAACTCATGTTAGTGATCATGCTGATAAGTTCCGGGCTGGCGGCCTTGGGTCTTAACCTGGTGCGGTCGTCGCTCGAGAACCTGATCCTGTCGTGCGTGTTCGAGGCCATCGTGAGCTGCACGGAGGCCGTGCTCTTCTGCGTCATCTGCGAGATCTTTCCCACTAAAGTCGCGGCGACGGCGATGGCGGTGACGGTGATGTGCGGCAGAGTGGGCGCCATCGTCGGCAACGTGGTGTTCGGCGCGCTGGTCGACGAGCACTGCATCGTGCCCATCTACATGTTCGGCACCCTCCTCATCACGAGCGGGCTGCTGTGCATCATCTTACCGAAGAGCACGCGACCGGAGCGGCCTCAGTGA
Protein
MEEHGESKDRVQRAPFETALHHAGYGCFQWLLLLSCGAVYAVCALSTTTLSFVLPATKNDFELSSSDKGRLTATPLIGMCVGSYFWGNLADARGRKIAIIGALLLDALAAFLSSVVQSFSGFLACRFFSGFGIIGATGLIFPYFGDFLSLRHRDVMLCRLEVFWTVGTILLPGIAFLIIPDKRFTLTAADADFQYTSWRVFVAACGVPSLLVVLLLLPFPESPRYLLHANRAEQALHVLRRIFVVNTKLDAKDFPVESMISAGDGDEEEVVSADNNGNETKAPLTWRQRWDAFWIRKKMLVTPPYIGLLVLCCFVDFGLMFSYYTLMMWFPDLFERFSQFASLYGDEPAGVCEVSLAVANATTDIPIQSTMSPVHDQVYIHTLVVALSCVPTALWVGLTINIVGKKLMLVIMLISSGLAALGLNLVRSSLENLILSCVFEAIVSCTEAVLFCVICEIFPTKVAATAMAVTVMCGRVGAIVGNVVFGALVDEHCIVPIYMFGTLLITSGLLCIILPKSTRPERPQ
Summary
Uniprot
A0A2W1BL90
H9JVB4
S4PAA6
A0A212F5C0
A0A2H1VVE4
A0A194Q3E4
+ More
A0A0N1PJE7 A0A2H1VUF3 A0A0L7L9G6 A0A139WMB8 A0A139WMH7 A0A1Y1KNN4 A0A1Y1KR70 D6WAI5 A0A1B0CZJ6 A0A2H1VMI9 H9IYU5 A0A212ESJ6 A0A194RSD6 B0WWU3 A0A1Q3FLS2 A0A194QBE5 A0A1A9WYM5 A0A0L0BTJ0 A0A1J1HTA4 A0A336MF81 A0A1Y1KTJ9 A0A3B0K2C3 A0A0C9R7R4 A0A084VV04 Q16N29 A0A1S4FVL6 A0A1B6CG08 K7IYL5 E2B477 A0A182J0Q5 A0A1B0FI37 A0A182VH93 E1ZV02 A0A026WFC5 A0A182HII0 E9J4Z0 A0A182KCL9 A0A182Q543 A0A2A4J710 A0A232FF43 N6U544 A0A158NM89 A0A182NAG2 A0A3B0KR71 A0A154PLN8 Q7QES5 A0A151JVX9 A0A195CWY6 A0A182RVU8 F4WHS8 A0A2H8TTN8 A0A1W4X2H9 A0A182YQT2 A0A182PU98 A0A1W4WRJ2 J9JQX5 A0A1W4X2A3 A0A151JPY9 A0A182W433 A0A0J7KHI2 A0A182FMD5 A0A0M8ZS97 A0A151X0M7 A0A195BRX6 A0A0L7RC69 A0A1B6H4Q3 A0A182WY91 A0A2R7W2U3 A0A3B0K296 A0A2S2QJC6 A0A182TDY5 A0A182S8V6 A0A0K8UP05 T1HJP6 A0A2A3EJA3 A0A146KZ19 W8C1C9 H9IW67 A0A2A4J951 A0A336MZY6 A0A034VRR2 A0A336N2T7 A0A336N2J8 S4PYD7 J9JQL3 A0A0N1I5D8 A0A1B6EF96 A0A1J1IPI5 A0A1L8DEU3 J9K5L1 A0A0A1WZ37 A0A212ESA5 A0A1I8PV12 A0A1L8DEM5
A0A0N1PJE7 A0A2H1VUF3 A0A0L7L9G6 A0A139WMB8 A0A139WMH7 A0A1Y1KNN4 A0A1Y1KR70 D6WAI5 A0A1B0CZJ6 A0A2H1VMI9 H9IYU5 A0A212ESJ6 A0A194RSD6 B0WWU3 A0A1Q3FLS2 A0A194QBE5 A0A1A9WYM5 A0A0L0BTJ0 A0A1J1HTA4 A0A336MF81 A0A1Y1KTJ9 A0A3B0K2C3 A0A0C9R7R4 A0A084VV04 Q16N29 A0A1S4FVL6 A0A1B6CG08 K7IYL5 E2B477 A0A182J0Q5 A0A1B0FI37 A0A182VH93 E1ZV02 A0A026WFC5 A0A182HII0 E9J4Z0 A0A182KCL9 A0A182Q543 A0A2A4J710 A0A232FF43 N6U544 A0A158NM89 A0A182NAG2 A0A3B0KR71 A0A154PLN8 Q7QES5 A0A151JVX9 A0A195CWY6 A0A182RVU8 F4WHS8 A0A2H8TTN8 A0A1W4X2H9 A0A182YQT2 A0A182PU98 A0A1W4WRJ2 J9JQX5 A0A1W4X2A3 A0A151JPY9 A0A182W433 A0A0J7KHI2 A0A182FMD5 A0A0M8ZS97 A0A151X0M7 A0A195BRX6 A0A0L7RC69 A0A1B6H4Q3 A0A182WY91 A0A2R7W2U3 A0A3B0K296 A0A2S2QJC6 A0A182TDY5 A0A182S8V6 A0A0K8UP05 T1HJP6 A0A2A3EJA3 A0A146KZ19 W8C1C9 H9IW67 A0A2A4J951 A0A336MZY6 A0A034VRR2 A0A336N2T7 A0A336N2J8 S4PYD7 J9JQL3 A0A0N1I5D8 A0A1B6EF96 A0A1J1IPI5 A0A1L8DEU3 J9K5L1 A0A0A1WZ37 A0A212ESA5 A0A1I8PV12 A0A1L8DEM5
Pubmed
EMBL
KZ149979
PZC75842.1
BABH01037426
GAIX01003339
JAA89221.1
AGBW02010212
+ More
OWR48923.1 ODYU01004504 SOQ44452.1 KQ459579 KPI99534.1 KQ459753 KPJ20153.1 ODYU01004503 SOQ44450.1 JTDY01002134 KOB72030.1 KQ971312 KYB29082.1 KYB29083.1 GEZM01080370 JAV62188.1 GEZM01080373 GEZM01080372 JAV62185.1 EEZ97996.1 AJVK01009607 ODYU01003368 SOQ42041.1 BABH01010760 AGBW02012778 OWR44449.1 KQ459984 KPJ18981.1 DS232152 EDS36204.1 GFDL01006577 JAV28468.1 KQ459220 KPJ02863.1 JRES01001365 KNC23336.1 CVRI01000020 CRK90612.1 UFQT01000487 SSX24708.1 GEZM01080371 JAV62187.1 OUUW01000014 SPP88384.1 GBYB01004070 JAG73837.1 ATLV01017139 KE525157 KFB41798.1 CH477841 EAT35757.1 GEDC01025023 JAS12275.1 GL445515 EFN89494.1 CCAG010001331 GL434335 EFN75000.1 KK107238 EZA54618.1 APCN01003972 GL768121 EFZ12128.1 AXCN02002022 NWSH01002966 PCG67203.1 NNAY01000295 OXU29386.1 APGK01042207 KB741002 ENN75761.1 ADTU01020150 SPP88386.1 KQ434960 KZC12677.1 AAAB01008846 EAA06762.5 KQ981688 KYN37931.1 KQ977171 KYN05178.1 GL888167 EGI66285.1 GFXV01005504 MBW17309.1 ABLF02017432 ABLF02017434 ABLF02032144 KQ978684 KYN29305.1 LBMM01007490 KMQ89702.1 KQ435911 KOX68926.1 KQ982610 KYQ53909.1 KQ976421 KYM89117.1 KQ414616 KOC68423.1 GECZ01000108 JAS69661.1 KK854234 PTY13511.1 SPP88387.1 GGMS01008652 MBY77855.1 GDHF01023897 GDHF01021039 GDHF01014115 GDHF01013948 GDHF01010407 GDHF01000263 JAI28417.1 JAI31275.1 JAI38199.1 JAI38366.1 JAI41907.1 JAI52051.1 ACPB03019799 KZ288226 PBC31873.1 GDHC01017015 JAQ01614.1 GAMC01011121 JAB95434.1 BABH01005928 BABH01005929 BABH01005930 NWSH01002480 PCG68208.1 UFQS01005126 UFQT01005126 SSX16721.1 SSX35912.1 GAKP01013803 GAKP01013802 GAKP01013801 JAC45149.1 UFQS01005213 UFQT01005213 SSX16759.1 SSX35949.1 UFQS01004952 UFQT01004952 SSX16632.1 SSX35829.1 GAIX01003468 JAA89092.1 ABLF02027891 KQ461068 KPJ09887.1 GEDC01000686 JAS36612.1 CVRI01000057 CRL02157.1 GFDF01009187 JAV04897.1 ABLF02018816 GBXI01017510 GBXI01010170 JAC96781.1 JAD04122.1 AGBW02012834 OWR44382.1 GFDF01009188 JAV04896.1
OWR48923.1 ODYU01004504 SOQ44452.1 KQ459579 KPI99534.1 KQ459753 KPJ20153.1 ODYU01004503 SOQ44450.1 JTDY01002134 KOB72030.1 KQ971312 KYB29082.1 KYB29083.1 GEZM01080370 JAV62188.1 GEZM01080373 GEZM01080372 JAV62185.1 EEZ97996.1 AJVK01009607 ODYU01003368 SOQ42041.1 BABH01010760 AGBW02012778 OWR44449.1 KQ459984 KPJ18981.1 DS232152 EDS36204.1 GFDL01006577 JAV28468.1 KQ459220 KPJ02863.1 JRES01001365 KNC23336.1 CVRI01000020 CRK90612.1 UFQT01000487 SSX24708.1 GEZM01080371 JAV62187.1 OUUW01000014 SPP88384.1 GBYB01004070 JAG73837.1 ATLV01017139 KE525157 KFB41798.1 CH477841 EAT35757.1 GEDC01025023 JAS12275.1 GL445515 EFN89494.1 CCAG010001331 GL434335 EFN75000.1 KK107238 EZA54618.1 APCN01003972 GL768121 EFZ12128.1 AXCN02002022 NWSH01002966 PCG67203.1 NNAY01000295 OXU29386.1 APGK01042207 KB741002 ENN75761.1 ADTU01020150 SPP88386.1 KQ434960 KZC12677.1 AAAB01008846 EAA06762.5 KQ981688 KYN37931.1 KQ977171 KYN05178.1 GL888167 EGI66285.1 GFXV01005504 MBW17309.1 ABLF02017432 ABLF02017434 ABLF02032144 KQ978684 KYN29305.1 LBMM01007490 KMQ89702.1 KQ435911 KOX68926.1 KQ982610 KYQ53909.1 KQ976421 KYM89117.1 KQ414616 KOC68423.1 GECZ01000108 JAS69661.1 KK854234 PTY13511.1 SPP88387.1 GGMS01008652 MBY77855.1 GDHF01023897 GDHF01021039 GDHF01014115 GDHF01013948 GDHF01010407 GDHF01000263 JAI28417.1 JAI31275.1 JAI38199.1 JAI38366.1 JAI41907.1 JAI52051.1 ACPB03019799 KZ288226 PBC31873.1 GDHC01017015 JAQ01614.1 GAMC01011121 JAB95434.1 BABH01005928 BABH01005929 BABH01005930 NWSH01002480 PCG68208.1 UFQS01005126 UFQT01005126 SSX16721.1 SSX35912.1 GAKP01013803 GAKP01013802 GAKP01013801 JAC45149.1 UFQS01005213 UFQT01005213 SSX16759.1 SSX35949.1 UFQS01004952 UFQT01004952 SSX16632.1 SSX35829.1 GAIX01003468 JAA89092.1 ABLF02027891 KQ461068 KPJ09887.1 GEDC01000686 JAS36612.1 CVRI01000057 CRL02157.1 GFDF01009187 JAV04897.1 ABLF02018816 GBXI01017510 GBXI01010170 JAC96781.1 JAD04122.1 AGBW02012834 OWR44382.1 GFDF01009188 JAV04896.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000037510
UP000007266
+ More
UP000092462 UP000002320 UP000091820 UP000037069 UP000183832 UP000268350 UP000030765 UP000008820 UP000002358 UP000008237 UP000075880 UP000092444 UP000075903 UP000000311 UP000053097 UP000075840 UP000075881 UP000075886 UP000218220 UP000215335 UP000019118 UP000005205 UP000075884 UP000076502 UP000007062 UP000078541 UP000078542 UP000075900 UP000007755 UP000192223 UP000076408 UP000075885 UP000007819 UP000078492 UP000075920 UP000036403 UP000069272 UP000053105 UP000075809 UP000078540 UP000053825 UP000076407 UP000075902 UP000075901 UP000015103 UP000242457 UP000095300
UP000092462 UP000002320 UP000091820 UP000037069 UP000183832 UP000268350 UP000030765 UP000008820 UP000002358 UP000008237 UP000075880 UP000092444 UP000075903 UP000000311 UP000053097 UP000075840 UP000075881 UP000075886 UP000218220 UP000215335 UP000019118 UP000005205 UP000075884 UP000076502 UP000007062 UP000078541 UP000078542 UP000075900 UP000007755 UP000192223 UP000076408 UP000075885 UP000007819 UP000078492 UP000075920 UP000036403 UP000069272 UP000053105 UP000075809 UP000078540 UP000053825 UP000076407 UP000075902 UP000075901 UP000015103 UP000242457 UP000095300
Interpro
CDD
ProteinModelPortal
A0A2W1BL90
H9JVB4
S4PAA6
A0A212F5C0
A0A2H1VVE4
A0A194Q3E4
+ More
A0A0N1PJE7 A0A2H1VUF3 A0A0L7L9G6 A0A139WMB8 A0A139WMH7 A0A1Y1KNN4 A0A1Y1KR70 D6WAI5 A0A1B0CZJ6 A0A2H1VMI9 H9IYU5 A0A212ESJ6 A0A194RSD6 B0WWU3 A0A1Q3FLS2 A0A194QBE5 A0A1A9WYM5 A0A0L0BTJ0 A0A1J1HTA4 A0A336MF81 A0A1Y1KTJ9 A0A3B0K2C3 A0A0C9R7R4 A0A084VV04 Q16N29 A0A1S4FVL6 A0A1B6CG08 K7IYL5 E2B477 A0A182J0Q5 A0A1B0FI37 A0A182VH93 E1ZV02 A0A026WFC5 A0A182HII0 E9J4Z0 A0A182KCL9 A0A182Q543 A0A2A4J710 A0A232FF43 N6U544 A0A158NM89 A0A182NAG2 A0A3B0KR71 A0A154PLN8 Q7QES5 A0A151JVX9 A0A195CWY6 A0A182RVU8 F4WHS8 A0A2H8TTN8 A0A1W4X2H9 A0A182YQT2 A0A182PU98 A0A1W4WRJ2 J9JQX5 A0A1W4X2A3 A0A151JPY9 A0A182W433 A0A0J7KHI2 A0A182FMD5 A0A0M8ZS97 A0A151X0M7 A0A195BRX6 A0A0L7RC69 A0A1B6H4Q3 A0A182WY91 A0A2R7W2U3 A0A3B0K296 A0A2S2QJC6 A0A182TDY5 A0A182S8V6 A0A0K8UP05 T1HJP6 A0A2A3EJA3 A0A146KZ19 W8C1C9 H9IW67 A0A2A4J951 A0A336MZY6 A0A034VRR2 A0A336N2T7 A0A336N2J8 S4PYD7 J9JQL3 A0A0N1I5D8 A0A1B6EF96 A0A1J1IPI5 A0A1L8DEU3 J9K5L1 A0A0A1WZ37 A0A212ESA5 A0A1I8PV12 A0A1L8DEM5
A0A0N1PJE7 A0A2H1VUF3 A0A0L7L9G6 A0A139WMB8 A0A139WMH7 A0A1Y1KNN4 A0A1Y1KR70 D6WAI5 A0A1B0CZJ6 A0A2H1VMI9 H9IYU5 A0A212ESJ6 A0A194RSD6 B0WWU3 A0A1Q3FLS2 A0A194QBE5 A0A1A9WYM5 A0A0L0BTJ0 A0A1J1HTA4 A0A336MF81 A0A1Y1KTJ9 A0A3B0K2C3 A0A0C9R7R4 A0A084VV04 Q16N29 A0A1S4FVL6 A0A1B6CG08 K7IYL5 E2B477 A0A182J0Q5 A0A1B0FI37 A0A182VH93 E1ZV02 A0A026WFC5 A0A182HII0 E9J4Z0 A0A182KCL9 A0A182Q543 A0A2A4J710 A0A232FF43 N6U544 A0A158NM89 A0A182NAG2 A0A3B0KR71 A0A154PLN8 Q7QES5 A0A151JVX9 A0A195CWY6 A0A182RVU8 F4WHS8 A0A2H8TTN8 A0A1W4X2H9 A0A182YQT2 A0A182PU98 A0A1W4WRJ2 J9JQX5 A0A1W4X2A3 A0A151JPY9 A0A182W433 A0A0J7KHI2 A0A182FMD5 A0A0M8ZS97 A0A151X0M7 A0A195BRX6 A0A0L7RC69 A0A1B6H4Q3 A0A182WY91 A0A2R7W2U3 A0A3B0K296 A0A2S2QJC6 A0A182TDY5 A0A182S8V6 A0A0K8UP05 T1HJP6 A0A2A3EJA3 A0A146KZ19 W8C1C9 H9IW67 A0A2A4J951 A0A336MZY6 A0A034VRR2 A0A336N2T7 A0A336N2J8 S4PYD7 J9JQL3 A0A0N1I5D8 A0A1B6EF96 A0A1J1IPI5 A0A1L8DEU3 J9K5L1 A0A0A1WZ37 A0A212ESA5 A0A1I8PV12 A0A1L8DEM5
PDB
4LDS
E-value=0.000162758,
Score=108
Ontologies
GO
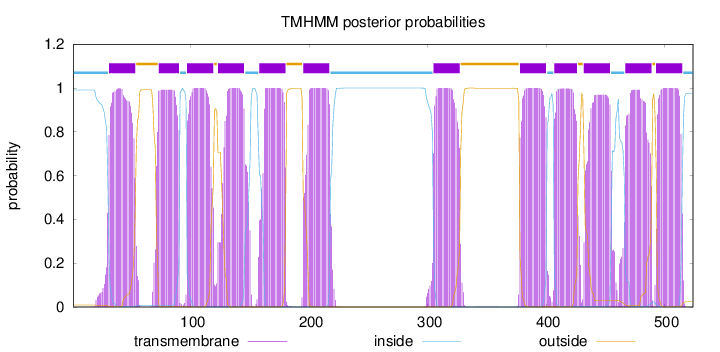
Topology
Length:
524
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
260.08345
Exp number, first 60 AAs:
23.08811
Total prob of N-in:
0.99067
POSSIBLE N-term signal
sequence
inside
1 - 30
TMhelix
31 - 53
outside
54 - 72
TMhelix
73 - 90
inside
91 - 96
TMhelix
97 - 119
outside
120 - 122
TMhelix
123 - 145
inside
146 - 157
TMhelix
158 - 180
outside
181 - 194
TMhelix
195 - 217
inside
218 - 304
TMhelix
305 - 327
outside
328 - 377
TMhelix
378 - 400
inside
401 - 406
TMhelix
407 - 426
outside
427 - 431
TMhelix
432 - 454
inside
455 - 466
TMhelix
467 - 489
outside
490 - 492
TMhelix
493 - 515
inside
516 - 524
Population Genetic Test Statistics
Pi
174.764628
Theta
168.732855
Tajima's D
0.254075
CLR
0.118353
CSRT
0.43972801359932
Interpretation
Uncertain
