Gene
KWMTBOMO03127
Pre Gene Modal
BGIBMGA005779
Annotation
PREDICTED:_zinc_transporter_2-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.535
Sequence
CDS
ATGTCATCACATTTAATTAACGAAAAATATCATAATGATACAAAAATAAATACATTGAACGCGTTCACTGATAAGAATTTAGTACCATTGAGTGGTAGATATAGATCGAAAAGTTTGTCTAGTGATAAGTTGAATAAAACAAACGCTGATCGTGCGGCTATATTGGAAGATGTCGAGGTCCAGTACATAACAGACAAGTCTGAGACACCTGGCTGTCTCGTCGGAAATAGAGATATGGTCAGCGGACAACTGAATATAAACGGGTACAATGATGGGCACGTATCGTACGGGACGGACAATCAGATCCGTGGGCGGAGAGTCATATTTTGTGTCCACGGGAACCCGTCGACGGGATGCTGTGCGGTCATCGAGACCAGCGCCGACGGCGATGACTCGGAGAAAAGGAACGGAAGCATCTCAGAGGCTGAAAGGCATTGTCACAGATCGCGAAACGAAGAGATAGACAAGAGAGCGAGAAGGAAACTCATCGTGGCCAGCGTGCTGTGCGTCATATTCATGATAGGGGAAATAGTAGGCGGGTACTTGTCCAACAGCTTAGCGATAGCGACCGACGCCGCGCACCTGCTCACCGACTTCGCCAGCTTCATGATATCCTTGTTCTCGCTATGGGTCGCTAACAGACCCGCCACCAGACGGATGTCGTTCGGGTGGTACCGCGCGGAGGTGATCGGCGCGCTGACGTCGGTGCTGCTCATCTGGGTGGTGACGGGCGTGCTGGTGTACCTCGCCGTGCAGCGGGTGCTCTACCAGGACTTCAACATCGACGCCACCGTCATGCTCATCACCTCCGCCGTCGGCGTCGCCGTCAACCTCGTCATGGGCCTGACTCTGCACCAGCACGGGCACTCGCACGGGGGAGGAGCCCATGGACACTCGCACGGACCCAGCAACCCCGTCCTCAACAACAGGGAGGTGGCGGGCGCGGAGGCGGGGGCCGGGGGGGCGGCGCGCGCGGAGAACATCAACGTGCGCGCCGCCTTCATCCACGTGCTCGGAGACTTCCTGCAGAGCTTCGGCGTGCTCGTCGCCGCCATCGTCATCTACTTCGAGCCGAGTTGGATCCTCATGGACCCGATCTGCACGTTCCTGTTTTCCATCCTGGTTCTCATCACGACCTTCAATATCATCAAAGACGCGCTTCTTGTGCTGATGGAGGGCTCCCCCCGAGGGCTGGACTTCCAGGAGGTGGCGGACACGTTCCTGGCGCTGCCGGGCGTGCTGCGCATCCACAACGTGCGCATGTGGGCGCTCACGCTCGACAAGCCCGCGCTCTCCGCGCACCTCGCCATCCGTAGCGGAGTCAGCCCGCAGAAAGTCTTGGAAGAGGCGACCCGTCTCGTCCACGAGAAGTACAACTTCTTCGAGATGACCCTCCAGATCGAAGAGTTCAGCGACGGCATGGAGCAGTGCAGCCAATGCAAGGTGCCCCAGGCCTGA
Protein
MSSHLINEKYHNDTKINTLNAFTDKNLVPLSGRYRSKSLSSDKLNKTNADRAAILEDVEVQYITDKSETPGCLVGNRDMVSGQLNINGYNDGHVSYGTDNQIRGRRVIFCVHGNPSTGCCAVIETSADGDDSEKRNGSISEAERHCHRSRNEEIDKRARRKLIVASVLCVIFMIGEIVGGYLSNSLAIATDAAHLLTDFASFMISLFSLWVANRPATRRMSFGWYRAEVIGALTSVLLIWVVTGVLVYLAVQRVLYQDFNIDATVMLITSAVGVAVNLVMGLTLHQHGHSHGGGAHGHSHGPSNPVLNNREVAGAEAGAGGAARAENINVRAAFIHVLGDFLQSFGVLVAAIVIYFEPSWILMDPICTFLFSILVLITTFNIIKDALLVLMEGSPRGLDFQEVADTFLALPGVLRIHNVRMWALTLDKPALSAHLAIRSGVSPQKVLEEATRLVHEKYNFFEMTLQIEEFSDGMEQCSQCKVPQA
Summary
Uniprot
A0A2W1BPI4
S4NNW6
A0A2A4JWG0
A0A0N1IE90
A0A212EYW4
A0A194PVM0
+ More
A0A2H1WLC0 A0A3L8DMU6 A0A026X3D2 A0A151I8Y3 A0A0L7RIU3 A0A1Y1LJA5 A0A1Y1LFL2 E2C512 A0A151WFV1 A0A1Y1LHS7 A0A088AHX8 A0A158NVB4 A0A310SBF8 N6TL29 A0A154P8B0 A0A0N0U358 A0A0J7L8I6 A0A2A3EBR5 A0A2J7QXR7 E2AEM6 A0A151J135 A0A195BU08 A0A1B6H9H4 A0A1B6IV94 A0A0P5WZ45 A0A146L1H4 A0A146KY03 A0A0P5NK62 A0A0K2TGL1 A0A336KV99 A0A146M9D9 A0A336MYR4 A0A084VKE9 A0A2I9LPZ2 A0A0P4ZUY3 T1E9Y5 A0A1B6G1C1 A0A195FXM8 A0A293N542 A0A1Z5KXN7 B4MVE8 W5MBF7 A0A0H3YKE4 I5AMN6 A0A1V9XAY6 K1R1V1 T2M9Y5 A0A1U8C809 G1SRW9 G1TUI5 A0A087X4V3 A0A1I8HLT5 A0A267DXD0 A0A267DDB9 A0A3P9AP17 A0A2R7W4F4 A0A1I8JKA6 A0A0L8FZN5 A0A0N5AI03 A0A3B5AXR7 A0A3B5KS11 A0A1W2WJS1 A0A094ZEU5 A0A0N4V2X9 A0A3Q0DQK8
A0A2H1WLC0 A0A3L8DMU6 A0A026X3D2 A0A151I8Y3 A0A0L7RIU3 A0A1Y1LJA5 A0A1Y1LFL2 E2C512 A0A151WFV1 A0A1Y1LHS7 A0A088AHX8 A0A158NVB4 A0A310SBF8 N6TL29 A0A154P8B0 A0A0N0U358 A0A0J7L8I6 A0A2A3EBR5 A0A2J7QXR7 E2AEM6 A0A151J135 A0A195BU08 A0A1B6H9H4 A0A1B6IV94 A0A0P5WZ45 A0A146L1H4 A0A146KY03 A0A0P5NK62 A0A0K2TGL1 A0A336KV99 A0A146M9D9 A0A336MYR4 A0A084VKE9 A0A2I9LPZ2 A0A0P4ZUY3 T1E9Y5 A0A1B6G1C1 A0A195FXM8 A0A293N542 A0A1Z5KXN7 B4MVE8 W5MBF7 A0A0H3YKE4 I5AMN6 A0A1V9XAY6 K1R1V1 T2M9Y5 A0A1U8C809 G1SRW9 G1TUI5 A0A087X4V3 A0A1I8HLT5 A0A267DXD0 A0A267DDB9 A0A3P9AP17 A0A2R7W4F4 A0A1I8JKA6 A0A0L8FZN5 A0A0N5AI03 A0A3B5AXR7 A0A3B5KS11 A0A1W2WJS1 A0A094ZEU5 A0A0N4V2X9 A0A3Q0DQK8
Pubmed
EMBL
KZ150027
PZC74786.1
GAIX01012109
JAA80451.1
NWSH01000548
PCG75730.1
+ More
KQ460970 KPJ10180.1 AGBW02011425 OWR46682.1 KQ459597 KPI95175.1 ODYU01009404 SOQ53807.1 QOIP01000006 RLU21249.1 KK107039 EZA61904.1 KQ978330 KYM95087.1 KQ414583 KOC70785.1 GEZM01057117 JAV72440.1 GEZM01057118 JAV72439.1 GL452714 EFN76966.1 KQ983203 KYQ46716.1 GEZM01057116 JAV72441.1 ADTU01027002 ADTU01027003 KQ775960 OAD52234.1 APGK01020367 KB740193 KB631602 ENN81139.1 ERL84537.1 KQ434826 KZC07448.1 KQ435969 KOX67852.1 LBMM01000270 KMR02239.1 KZ288310 PBC28649.1 NEVH01009372 PNF33371.1 GL438870 EFN68101.1 KQ980575 KYN15496.1 KQ976417 KYM89904.1 GECU01036331 JAS71375.1 GECU01016842 JAS90864.1 GDIP01079842 JAM23873.1 GDHC01016820 JAQ01809.1 GDHC01017930 JAQ00699.1 GDIQ01141140 JAL10586.1 HACA01007817 HACA01007818 CDW25178.1 UFQS01001002 UFQT01001002 SSX08470.1 SSX28404.1 GDHC01002266 JAQ16363.1 UFQS01001562 UFQT01001562 SSX11451.1 SSX31018.1 ATLV01014190 ATLV01014191 ATLV01014192 ATLV01014193 ATLV01014194 KE524948 KFB38443.1 GFWZ01000477 MBW20467.1 GDIP01207688 JAJ15714.1 GAMD01000887 JAB00704.1 GECZ01013549 JAS56220.1 KQ981193 KYN45181.1 GFWV01022581 MAA47308.1 GFJQ02007150 JAV99819.1 CH963857 EDW76493.2 AHAT01009594 KT163678 AKN21628.1 CH379060 EIM52221.1 MNPL01017151 OQR70561.1 JH816438 EKC35075.1 HAAD01002712 CDG68944.1 AAGW02062867 AYCK01018097 NIVC01003111 PAA53239.1 NIVC01004502 PAA47290.1 KK854274 PTY14091.1 KQ425015 KOF70123.1 KL250523 KGB32935.1 UXUI01007760 VDD89337.1
KQ460970 KPJ10180.1 AGBW02011425 OWR46682.1 KQ459597 KPI95175.1 ODYU01009404 SOQ53807.1 QOIP01000006 RLU21249.1 KK107039 EZA61904.1 KQ978330 KYM95087.1 KQ414583 KOC70785.1 GEZM01057117 JAV72440.1 GEZM01057118 JAV72439.1 GL452714 EFN76966.1 KQ983203 KYQ46716.1 GEZM01057116 JAV72441.1 ADTU01027002 ADTU01027003 KQ775960 OAD52234.1 APGK01020367 KB740193 KB631602 ENN81139.1 ERL84537.1 KQ434826 KZC07448.1 KQ435969 KOX67852.1 LBMM01000270 KMR02239.1 KZ288310 PBC28649.1 NEVH01009372 PNF33371.1 GL438870 EFN68101.1 KQ980575 KYN15496.1 KQ976417 KYM89904.1 GECU01036331 JAS71375.1 GECU01016842 JAS90864.1 GDIP01079842 JAM23873.1 GDHC01016820 JAQ01809.1 GDHC01017930 JAQ00699.1 GDIQ01141140 JAL10586.1 HACA01007817 HACA01007818 CDW25178.1 UFQS01001002 UFQT01001002 SSX08470.1 SSX28404.1 GDHC01002266 JAQ16363.1 UFQS01001562 UFQT01001562 SSX11451.1 SSX31018.1 ATLV01014190 ATLV01014191 ATLV01014192 ATLV01014193 ATLV01014194 KE524948 KFB38443.1 GFWZ01000477 MBW20467.1 GDIP01207688 JAJ15714.1 GAMD01000887 JAB00704.1 GECZ01013549 JAS56220.1 KQ981193 KYN45181.1 GFWV01022581 MAA47308.1 GFJQ02007150 JAV99819.1 CH963857 EDW76493.2 AHAT01009594 KT163678 AKN21628.1 CH379060 EIM52221.1 MNPL01017151 OQR70561.1 JH816438 EKC35075.1 HAAD01002712 CDG68944.1 AAGW02062867 AYCK01018097 NIVC01003111 PAA53239.1 NIVC01004502 PAA47290.1 KK854274 PTY14091.1 KQ425015 KOF70123.1 KL250523 KGB32935.1 UXUI01007760 VDD89337.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000279307
UP000053097
+ More
UP000078542 UP000053825 UP000008237 UP000075809 UP000005203 UP000005205 UP000019118 UP000030742 UP000076502 UP000053105 UP000036403 UP000242457 UP000235965 UP000000311 UP000078492 UP000078540 UP000030765 UP000078541 UP000007798 UP000018468 UP000001819 UP000192247 UP000005408 UP000189706 UP000001811 UP000028760 UP000095280 UP000215902 UP000265140 UP000053454 UP000046393 UP000261400 UP000005226 UP000038041 UP000274131 UP000189704
UP000078542 UP000053825 UP000008237 UP000075809 UP000005203 UP000005205 UP000019118 UP000030742 UP000076502 UP000053105 UP000036403 UP000242457 UP000235965 UP000000311 UP000078492 UP000078540 UP000030765 UP000078541 UP000007798 UP000018468 UP000001819 UP000192247 UP000005408 UP000189706 UP000001811 UP000028760 UP000095280 UP000215902 UP000265140 UP000053454 UP000046393 UP000261400 UP000005226 UP000038041 UP000274131 UP000189704
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BPI4
S4NNW6
A0A2A4JWG0
A0A0N1IE90
A0A212EYW4
A0A194PVM0
+ More
A0A2H1WLC0 A0A3L8DMU6 A0A026X3D2 A0A151I8Y3 A0A0L7RIU3 A0A1Y1LJA5 A0A1Y1LFL2 E2C512 A0A151WFV1 A0A1Y1LHS7 A0A088AHX8 A0A158NVB4 A0A310SBF8 N6TL29 A0A154P8B0 A0A0N0U358 A0A0J7L8I6 A0A2A3EBR5 A0A2J7QXR7 E2AEM6 A0A151J135 A0A195BU08 A0A1B6H9H4 A0A1B6IV94 A0A0P5WZ45 A0A146L1H4 A0A146KY03 A0A0P5NK62 A0A0K2TGL1 A0A336KV99 A0A146M9D9 A0A336MYR4 A0A084VKE9 A0A2I9LPZ2 A0A0P4ZUY3 T1E9Y5 A0A1B6G1C1 A0A195FXM8 A0A293N542 A0A1Z5KXN7 B4MVE8 W5MBF7 A0A0H3YKE4 I5AMN6 A0A1V9XAY6 K1R1V1 T2M9Y5 A0A1U8C809 G1SRW9 G1TUI5 A0A087X4V3 A0A1I8HLT5 A0A267DXD0 A0A267DDB9 A0A3P9AP17 A0A2R7W4F4 A0A1I8JKA6 A0A0L8FZN5 A0A0N5AI03 A0A3B5AXR7 A0A3B5KS11 A0A1W2WJS1 A0A094ZEU5 A0A0N4V2X9 A0A3Q0DQK8
A0A2H1WLC0 A0A3L8DMU6 A0A026X3D2 A0A151I8Y3 A0A0L7RIU3 A0A1Y1LJA5 A0A1Y1LFL2 E2C512 A0A151WFV1 A0A1Y1LHS7 A0A088AHX8 A0A158NVB4 A0A310SBF8 N6TL29 A0A154P8B0 A0A0N0U358 A0A0J7L8I6 A0A2A3EBR5 A0A2J7QXR7 E2AEM6 A0A151J135 A0A195BU08 A0A1B6H9H4 A0A1B6IV94 A0A0P5WZ45 A0A146L1H4 A0A146KY03 A0A0P5NK62 A0A0K2TGL1 A0A336KV99 A0A146M9D9 A0A336MYR4 A0A084VKE9 A0A2I9LPZ2 A0A0P4ZUY3 T1E9Y5 A0A1B6G1C1 A0A195FXM8 A0A293N542 A0A1Z5KXN7 B4MVE8 W5MBF7 A0A0H3YKE4 I5AMN6 A0A1V9XAY6 K1R1V1 T2M9Y5 A0A1U8C809 G1SRW9 G1TUI5 A0A087X4V3 A0A1I8HLT5 A0A267DXD0 A0A267DDB9 A0A3P9AP17 A0A2R7W4F4 A0A1I8JKA6 A0A0L8FZN5 A0A0N5AI03 A0A3B5AXR7 A0A3B5KS11 A0A1W2WJS1 A0A094ZEU5 A0A0N4V2X9 A0A3Q0DQK8
Ontologies
GO
PANTHER
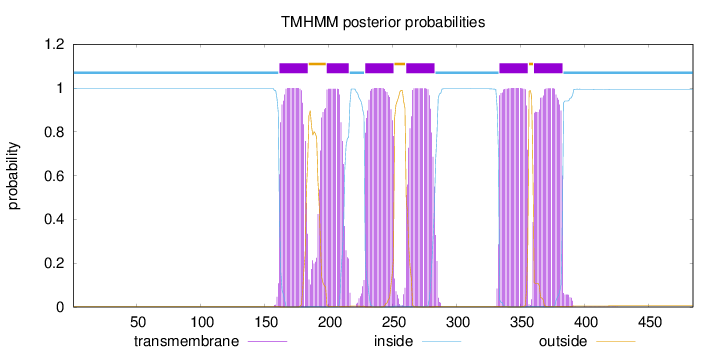
Topology
Length:
485
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
130.70027
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99834
inside
1 - 161
TMhelix
162 - 184
outside
185 - 198
TMhelix
199 - 216
inside
217 - 228
TMhelix
229 - 251
outside
252 - 260
TMhelix
261 - 283
inside
284 - 333
TMhelix
334 - 356
outside
357 - 360
TMhelix
361 - 383
inside
384 - 485
Population Genetic Test Statistics
Pi
2.519487
Theta
20.385644
Tajima's D
-0.895076
CLR
1.577081
CSRT
0.15829208539573
Interpretation
Uncertain
