Gene
KWMTBOMO03126
Pre Gene Modal
BGIBMGA005778
Annotation
Amine_oxidase_(Flavin-containing)_[Operophtera_brumata]
Full name
Amine oxidase
Location in the cell
Nuclear Reliability : 2.301
Sequence
CDS
ATGGTACAGAAACTCGATGAAACTCATAAGAAGAGTGTTAAAGCTGACGTAATTATTTTAGGATGCAGTTTATCAGGGATCGTGGCCGCTCACAAGCTAAAGAGGAGGTTCGGAGATTCTATGGATATTGTCGTGTTAGACCTGGCGGGACAGACACAAAGCTATTCCAAGTATAATGTTGTCTTCGAAGATGTAGAAAAGGACAGCCAGGATATCATAACCGAGCCGGATTTTCAAGGAACCGCTAAGCAAATGATTGAGAATGTTGCGCGGTTTTATCTAGCTAAATATGCCAAAGAATTCCACGTGCCCCTTCCAGACGTCATAATAGCCCCGGAAAGAGTCAGGACTAGGTTGACCAAGCTGTTCCAGCACCAAAACGGTAACACTGTAGAATGTACCAATGACTTCCACGAGTTCGACTATCTCAGCGTGATGGAGAGGTTCGAGATTAACCAATACCAGAACATGTTAGATCAGAGCATGAAGGACTTATTTCAGAGACACAAAGTAGATTTGCAAGCAGAAAGAAATAGGTTAAGATACTACGATCAGACGTGTATGGAAAGTCATATACGGGGGGCATTGTTGTTTCCAACTTCTAAAGAAATAATGAGATCTACCGTTAAGCTGGTCTGTGGCGCACCGGCTAATTCTGTGTCTGTGTTATTCTATCTTCATCAATGTTACAGGACCGGCAGTACTAAAAACCATTTAGATGGAAATAATACAAAGTTTAGGGAGAAGACTTTGGGTCACTGTCGGAAACGTCTCCAGAAAAAACTGCAGCAGAGCATCGCTGACATAACTCTGTCCGCTAAATCTATTAAGGAAATAAGGACATACTCTGATGAACAAGTTATTCTGGAAACAATAAAAGGCGAAACAGATTATGTGTGTAATTTGTTGGCGATGGCATTAAAACCTGACCAGCTCGACAGCATTCGAGTTGAGGATGAGCTGCTGGCGGTAAATTTGGCTAATAAGATTCGCGCCATGATTCCGGGTCGTGCCAGGAGGTTTGTTATACAATATGAAGATAACTTTTGGACGAAATTGGGCTACAGCGGTGACATTCTCAGCATACGTGGTCCAATCATCTGGGCAATGGAGAAACCAGAAATGTCTACGACTGGTAGCTTAGAGAAATATCCCTCACTGATCGGGTATTTGATGGTCCGTGATGAGAGTGACGGGGACAGTAAAGAAGCCGTCATAGAACAACTTATAAAACTATTCGGAAATAACGCAAGTTCCCCAGTGAACTACAAAGAGACAAACATAGCGGATTTGTTCATACCTAGGTGTGGAGATTATGTAGCACTTAAAGAGCTAACAGGCGAAGGGAGTCCGAAACACTTAGAGTGGGGGACGTTGGACATATTCGCCGATGGAGACGTTGCTGCTGCTCTGGAAGCTGGGCACACTGCCTACCTTAATCTGCTGACCAGCCTCCGTCCGCAAGCTCAGACATACGAGGATCGGAGTACGGCAGATTGGCCACGGTTTCTGGAAGATAGCCCTGTTCAAAGGTGGTATTCCCACGTAAATTTTGCTAATGGTATACATTTACTGGTGTACTCTGCTGCTGCGTATTACGGCTTACGTGTTGTGAGGTCTTATTTCCGGAAATAA
Protein
MVQKLDETHKKSVKADVIILGCSLSGIVAAHKLKRRFGDSMDIVVLDLAGQTQSYSKYNVVFEDVEKDSQDIITEPDFQGTAKQMIENVARFYLAKYAKEFHVPLPDVIIAPERVRTRLTKLFQHQNGNTVECTNDFHEFDYLSVMERFEINQYQNMLDQSMKDLFQRHKVDLQAERNRLRYYDQTCMESHIRGALLFPTSKEIMRSTVKLVCGAPANSVSVLFYLHQCYRTGSTKNHLDGNNTKFREKTLGHCRKRLQKKLQQSIADITLSAKSIKEIRTYSDEQVILETIKGETDYVCNLLAMALKPDQLDSIRVEDELLAVNLANKIRAMIPGRARRFVIQYEDNFWTKLGYSGDILSIRGPIIWAMEKPEMSTTGSLEKYPSLIGYLMVRDESDGDSKEAVIEQLIKLFGNNASSPVNYKETNIADLFIPRCGDYVALKELTGEGSPKHLEWGTLDIFADGDVAAALEAGHTAYLNLLTSLRPQAQTYEDRSTADWPRFLEDSPVQRWYSHVNFANGIHLLVYSAAAYYGLRVVRSYFRK
Summary
Cofactor
FAD
Similarity
Belongs to the flavin monoamine oxidase family.
Belongs to the ubiquitin-conjugating enzyme family.
Belongs to the ubiquitin-conjugating enzyme family.
Uniprot
H9J8D6
A0A0L7LI09
A0A2W1BMD8
A0A2H1VNU8
A0A2A4JUP7
A0A194PR00
+ More
A0A0N1PIG1 A0A212EZ07 W8CAZ6 A0A0A1XMS9 A0A139WAP1 A0A0K8UBZ3 B4G4F3 A0A1B0FL80 Q298P9 A0A2T5MC95 A0A183V872 A0A2J7RFP4 A0A1I8QA19 A0A0B2VW87 A0A368G2B3 A0A2A6C1R2 A0A3M6U997 A0A0F8B5V5 A0A2B4SVE0 K1QUT5 A0A1A9Y939 A0A1S3LB79 A0A2C9L605 A0A1S3LB49 A0A2T0PTG1 A0A1B0B6T4 A0A1S3L9V1 A0A3B4UCG2 A0A1S3LB54 W4XCR9 A0A0N8DFD6 A0A1N6ZIN9 A0A3E0C1A1 A0A0P6HDM0 A0A0P5N0X4 A0A3B3Q4Z2 A0A0R4IEI2 H3ET01 D3T0G7 H2SGI9 F1A178 A0A3B4WJ43 A0A1S3L9T0 A0A1M7R2B6 A0A3B4WBJ4 A0A1S3LB89 W5NGU4 A0A1W4YEG6 A0A1S3L9W1
A0A0N1PIG1 A0A212EZ07 W8CAZ6 A0A0A1XMS9 A0A139WAP1 A0A0K8UBZ3 B4G4F3 A0A1B0FL80 Q298P9 A0A2T5MC95 A0A183V872 A0A2J7RFP4 A0A1I8QA19 A0A0B2VW87 A0A368G2B3 A0A2A6C1R2 A0A3M6U997 A0A0F8B5V5 A0A2B4SVE0 K1QUT5 A0A1A9Y939 A0A1S3LB79 A0A2C9L605 A0A1S3LB49 A0A2T0PTG1 A0A1B0B6T4 A0A1S3L9V1 A0A3B4UCG2 A0A1S3LB54 W4XCR9 A0A0N8DFD6 A0A1N6ZIN9 A0A3E0C1A1 A0A0P6HDM0 A0A0P5N0X4 A0A3B3Q4Z2 A0A0R4IEI2 H3ET01 D3T0G7 H2SGI9 F1A178 A0A3B4WJ43 A0A1S3L9T0 A0A1M7R2B6 A0A3B4WBJ4 A0A1S3LB89 W5NGU4 A0A1W4YEG6 A0A1S3L9W1
EC Number
1.4.3.-
Pubmed
EMBL
BABH01027310
JTDY01000996
KOB75163.1
KZ150027
PZC74785.1
ODYU01003559
+ More
SOQ42478.1 NWSH01000548 PCG75731.1 KQ459597 KPI95174.1 KQ460970 KPJ10182.1 AGBW02011425 OWR46684.1 GAMC01000909 JAC05647.1 GBXI01002102 JAD12190.1 KQ971381 KYB24973.1 GDHF01028127 JAI24187.1 CH479179 EDW24501.1 CCAG010015535 CM000070 EAL27907.2 QANS01000007 PTU30193.1 UYWY01024018 VDM48263.1 NEVH01004410 PNF39652.1 JPKZ01000843 KHN85210.1 JOJR01000471 RCN37409.1 ABKE03000063 PDM72037.1 RCHS01001996 RMX50250.1 KQ041558 KKF25500.1 LSMT01000008 PFX33851.1 JH818947 EKC40562.1 PVZB01000035 PRX92184.1 JXJN01009220 AAGJ04120194 AAGJ04120195 GDIP01039159 LRGB01002100 JAM64556.1 KZS09220.1 CP019327 FTNP01000001 APX95340.1 SIR26657.1 QUNA01000002 REG49215.1 GDIQ01020734 JAN74003.1 GDIQ01148682 JAL03044.1 BX005125 CABZ01059629 CR925824 CP001932 AOHS01000024 ADD06446.1 ELY31667.1 GL871363 EGC30059.1 FRCS01000006 SHN39096.1 AHAT01008591
SOQ42478.1 NWSH01000548 PCG75731.1 KQ459597 KPI95174.1 KQ460970 KPJ10182.1 AGBW02011425 OWR46684.1 GAMC01000909 JAC05647.1 GBXI01002102 JAD12190.1 KQ971381 KYB24973.1 GDHF01028127 JAI24187.1 CH479179 EDW24501.1 CCAG010015535 CM000070 EAL27907.2 QANS01000007 PTU30193.1 UYWY01024018 VDM48263.1 NEVH01004410 PNF39652.1 JPKZ01000843 KHN85210.1 JOJR01000471 RCN37409.1 ABKE03000063 PDM72037.1 RCHS01001996 RMX50250.1 KQ041558 KKF25500.1 LSMT01000008 PFX33851.1 JH818947 EKC40562.1 PVZB01000035 PRX92184.1 JXJN01009220 AAGJ04120194 AAGJ04120195 GDIP01039159 LRGB01002100 JAM64556.1 KZS09220.1 CP019327 FTNP01000001 APX95340.1 SIR26657.1 QUNA01000002 REG49215.1 GDIQ01020734 JAN74003.1 GDIQ01148682 JAL03044.1 BX005125 CABZ01059629 CR925824 CP001932 AOHS01000024 ADD06446.1 ELY31667.1 GL871363 EGC30059.1 FRCS01000006 SHN39096.1 AHAT01008591
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000007266 UP000008744 UP000092444 UP000001819 UP000244248 UP000050794 UP000267007 UP000235965 UP000095300 UP000031036 UP000252519 UP000232936 UP000275408 UP000225706 UP000005408 UP000092443 UP000087266 UP000076420 UP000237679 UP000092460 UP000261420 UP000007110 UP000076858 UP000185687 UP000187321 UP000256904 UP000261540 UP000000437 UP000005239 UP000001879 UP000011543 UP000005226 UP000001064 UP000261360 UP000184440 UP000018468 UP000192224
UP000007266 UP000008744 UP000092444 UP000001819 UP000244248 UP000050794 UP000267007 UP000235965 UP000095300 UP000031036 UP000252519 UP000232936 UP000275408 UP000225706 UP000005408 UP000092443 UP000087266 UP000076420 UP000237679 UP000092460 UP000261420 UP000007110 UP000076858 UP000185687 UP000187321 UP000256904 UP000261540 UP000000437 UP000005239 UP000001879 UP000011543 UP000005226 UP000001064 UP000261360 UP000184440 UP000018468 UP000192224
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J8D6
A0A0L7LI09
A0A2W1BMD8
A0A2H1VNU8
A0A2A4JUP7
A0A194PR00
+ More
A0A0N1PIG1 A0A212EZ07 W8CAZ6 A0A0A1XMS9 A0A139WAP1 A0A0K8UBZ3 B4G4F3 A0A1B0FL80 Q298P9 A0A2T5MC95 A0A183V872 A0A2J7RFP4 A0A1I8QA19 A0A0B2VW87 A0A368G2B3 A0A2A6C1R2 A0A3M6U997 A0A0F8B5V5 A0A2B4SVE0 K1QUT5 A0A1A9Y939 A0A1S3LB79 A0A2C9L605 A0A1S3LB49 A0A2T0PTG1 A0A1B0B6T4 A0A1S3L9V1 A0A3B4UCG2 A0A1S3LB54 W4XCR9 A0A0N8DFD6 A0A1N6ZIN9 A0A3E0C1A1 A0A0P6HDM0 A0A0P5N0X4 A0A3B3Q4Z2 A0A0R4IEI2 H3ET01 D3T0G7 H2SGI9 F1A178 A0A3B4WJ43 A0A1S3L9T0 A0A1M7R2B6 A0A3B4WBJ4 A0A1S3LB89 W5NGU4 A0A1W4YEG6 A0A1S3L9W1
A0A0N1PIG1 A0A212EZ07 W8CAZ6 A0A0A1XMS9 A0A139WAP1 A0A0K8UBZ3 B4G4F3 A0A1B0FL80 Q298P9 A0A2T5MC95 A0A183V872 A0A2J7RFP4 A0A1I8QA19 A0A0B2VW87 A0A368G2B3 A0A2A6C1R2 A0A3M6U997 A0A0F8B5V5 A0A2B4SVE0 K1QUT5 A0A1A9Y939 A0A1S3LB79 A0A2C9L605 A0A1S3LB49 A0A2T0PTG1 A0A1B0B6T4 A0A1S3L9V1 A0A3B4UCG2 A0A1S3LB54 W4XCR9 A0A0N8DFD6 A0A1N6ZIN9 A0A3E0C1A1 A0A0P6HDM0 A0A0P5N0X4 A0A3B3Q4Z2 A0A0R4IEI2 H3ET01 D3T0G7 H2SGI9 F1A178 A0A3B4WJ43 A0A1S3L9T0 A0A1M7R2B6 A0A3B4WBJ4 A0A1S3LB89 W5NGU4 A0A1W4YEG6 A0A1S3L9W1
PDB
1O5W
E-value=8.68287e-05,
Score=111
Ontologies
PATHWAY
00260
Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00350 Tyrosine metabolism - Bombyx mori (domestic silkworm)
00360 Phenylalanine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00350 Tyrosine metabolism - Bombyx mori (domestic silkworm)
00360 Phenylalanine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
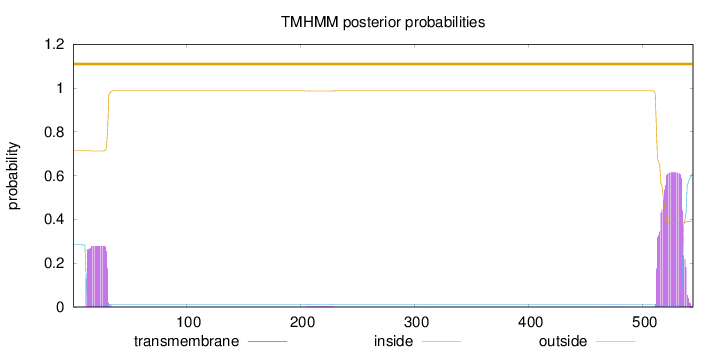
Length:
544
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
18.74274
Exp number, first 60 AAs:
5.27575
Total prob of N-in:
0.28386
outside
1 - 544
Population Genetic Test Statistics
Pi
5.818775
Theta
7.105001
Tajima's D
-0.567209
CLR
0.003831
CSRT
0.226638668066597
Interpretation
Uncertain
