Gene
KWMTBOMO03125
Annotation
reverse_ribonuclease_integrase?_partial_[Lasius_niger]
Location in the cell
Mitochondrial Reliability : 2.139
Sequence
CDS
ATGCTCCAGCTTGATGTAGTTGAGCCTTGTGAGAGTGCCTGGCAATCCCCGGTATTGTTAGTCACTAAAAAAGATGGTCAGCCACGGTTTTGCTTAGATAGTAGGAAGCTAAACTCTGTTACGAAACGTGACGCTTATAACTTACCTTACATATCGGAGATATTGGACAACTTGCGAGACGCGCGCTATCTTAGCAGTATTGACTTATCAAAGGCTTTCTGGCAATTACCAATAGCACTCGAAGATAGGGACAAGACGGCATTTTACGTTCCGGGACGAGGGACGTTACGGTTCAAGACTACACCGTTTGGTTTGACCAATGCGCCTTCTACGCAACAGCGGTTAGTCGATTTATTGTTTGGCAACATGGATCACAAGGCGTTCGCATATTTAGATGATGTGATAATAGTAAGTAGCACGTTTGATGAGCATGTTAGTCTTCTTCTTCGCGTGTTAGACAAACTGCGCCATGCTAATCTAACAATTAATTTAGAAAAGTGTGAATTTTTTCGAAGTAAACTTAAATATTTAGGCTATGTGGTCGACGCGAATGGTTTGCGGACTGATCCGGATAAAGTTACAGCCATTCTAAATTACCCAACTCCTACATGTCGTAAGGACTTGAAACGGTTCCTAGGCACGGCAACATGGTACCGGAGATTTGTCCCAAACTTTAGCACTATCGCAGGGCCGCTTAATAGGTTGACATCGAACAAAAAATCAACACATCCCTTCCAGTGGACAGATGAGGCCGACACTGCGTTCAAAAAACTTAAGGAATCTTTGGTCTCGGCACCTGTTCTCTCCTGCCCAAATTATGATCTGCCTTTCGAAGTCCATACAGATGCAAGTAACTATGGGATAGGTGGTATGCTCACACAAACTATTGACGGGAAAGAGCACCCTATAGCGTATATGAGTAAGTCCTTATCGGGGGCCGAAAAGAATTATAGCATTACCGAACGTGAGACCTTAGCTGTTCTCGTGGCTCTCGAGCACTGGCGTTGCTATCTGGAGAACGGAAAGACATTCACGGTTTACACAGACCACAGCGCACTTAAGTGGTTCCTTTCCTTGAACAATCCTACGGGTAGGCTTGCACGTTGGGGAGTTCGATTATCGTCGTTTGATTTTGAAATCAAACACCGCCGCGGTGTAGACAATGTGATTCCCGACGCTTTGTCTAGAGCTGTCCCGGTCACTACTATCACTACTGCTAAATCACTCACGACTTCTAGTGATGATTGAAAGCCTTTTGAATTTAATGTAGGAGATACTGTTTATAAACGCGCTTACGTCCTGAGTGACAAAGATAAGTATTTTAGTAAGAAGTTAGCACCGAAATATATCAAGTGCAAAGTTGTCGAAAAACTCTCGCCACTTGTCTACGTCCTAGAAGACATGTCGGGTAAAAACATAGGCACTTGGCATATTAAAGACTTGAAAATATTAGGACAACACTCATAG
Protein
MLQLDVVEPCESAWQSPVLLVTKKDGQPRFCLDSRKLNSVTKRDAYNLPYISEILDNLRDARYLSSIDLSKAFWQLPIALEDRDKTAFYVPGRGTLRFKTTPFGLTNAPSTQQRLVDLLFGNMDHKAFAYLDDVIIVSSTFDEHVSLLLRVLDKLRHANLTINLEKCEFFRSKLKYLGYVVDANGLRTDPDKVTAILNYPTPTCRKDLKRFLGTATWYRRFVPNFSTIAGPLNRLTSNKKSTHPFQWTDEADTAFKKLKESLVSAPVLSCPNYDLPFEVHTDASNYGIGGMLTQTIDGKEHPIAYMSKSLSGAEKNYSITERETLAVLVALEHWRCYLENGKTFTVYTDHSALKWFLSLNNPTGRLARWGVRLSSFDFEIKHRRGVDNVIPDALSRAVPVTTITTAKSLTTSSDDXKPFEFNVGDTVYKRAYVLSDKDKYFSKKLAPKYIKCKVVEKLSPLVYVLEDMSGKNIGTWHIKDLKILGQHS
Summary
Uniprot
A0A1Y1MJD1
V5H0H7
A0A146LTK8
A0A1Y1MED9
A0A1Y1NMV4
A0A1Y1JW20
+ More
A0A1Y1K180 A0A0A9XMW3 A0A1B6MMN8 A0A0J7JZY0 A0A1Y1JUB3 A0A1Y1K0I6 A0A0J7K1V9 A0A1Y1JVH7 W8AD09 A0A1B6LNQ5 A0A1Y1LZS5 A0A1Y1MVI4 A0A224XH29 A0A023EY78 A0A224XHI1 A0A0J7KA76 A0A1Y1MIJ6 V5GNV6 A0A224XDN8 A0A224XHE2 A0A1Y1L0V4 A0A0C9QRC9 A0A1B6IL28 A0A0J7K794 A0A0A9YUQ6 A0A139WG74 A0A0J7K9T8 A0A0J7KA70 A0A023EXS9 A0A1Y1NGJ5 A0A0J7K8R1 A0A0J7KPJ7 A0A224X7A2 A0A182GM87 A0A0J7KE06 A0A0J7KDB6 A0A139W8U5 V5I839 A0A1Y1KNF8 A0A1Y1MZ56 A0A0J7K8Q7 A0A224X632 A0A0J7K4Z4 A0A1Y1MWM5 A0A1Y1N3D6 A0A0A9Z8Q1 A0A0A9VXA9 A0A0J7K489 A0A0A9W1A6 A0A146KR86 A0A0J7K1N1 A0A146LKA7 A0A0V0G672 A0A0J7K441 A0A1Y1LV33 A0A146LWQ9 A0A224X6V5 A0A139W930 A0A0R1E8A2 A0A0J7K1I1 A0A023EY81 K7JCB2 W8B7U6 A0A0C9PJT1 K7JB64 A0A0J7K3V6 A0A087SUZ9 A0A087T3I4 A0A182H681 A0A1Z5KVF1 Q9XZR7 A1BPH1 A0A0J7K3C0 A0A0S7KB76 A0A087T293 A0A0J7K313 A0A0K3CWF3 A0A3B3IK80 A0A1W7R6D0 A0A0S7KAF1 L7MMB3 A0A0P6ATU6 A0A0J7KTY6 A0A1B6JW27 A0A147BST7 A0A1B6E3H6 A0A1L8DJ04 W4Y9K9 K7JCB3 L7LU37 A0A2G8JTJ5 A0A3B1JJM2 A0A2G8KXK5
A0A1Y1K180 A0A0A9XMW3 A0A1B6MMN8 A0A0J7JZY0 A0A1Y1JUB3 A0A1Y1K0I6 A0A0J7K1V9 A0A1Y1JVH7 W8AD09 A0A1B6LNQ5 A0A1Y1LZS5 A0A1Y1MVI4 A0A224XH29 A0A023EY78 A0A224XHI1 A0A0J7KA76 A0A1Y1MIJ6 V5GNV6 A0A224XDN8 A0A224XHE2 A0A1Y1L0V4 A0A0C9QRC9 A0A1B6IL28 A0A0J7K794 A0A0A9YUQ6 A0A139WG74 A0A0J7K9T8 A0A0J7KA70 A0A023EXS9 A0A1Y1NGJ5 A0A0J7K8R1 A0A0J7KPJ7 A0A224X7A2 A0A182GM87 A0A0J7KE06 A0A0J7KDB6 A0A139W8U5 V5I839 A0A1Y1KNF8 A0A1Y1MZ56 A0A0J7K8Q7 A0A224X632 A0A0J7K4Z4 A0A1Y1MWM5 A0A1Y1N3D6 A0A0A9Z8Q1 A0A0A9VXA9 A0A0J7K489 A0A0A9W1A6 A0A146KR86 A0A0J7K1N1 A0A146LKA7 A0A0V0G672 A0A0J7K441 A0A1Y1LV33 A0A146LWQ9 A0A224X6V5 A0A139W930 A0A0R1E8A2 A0A0J7K1I1 A0A023EY81 K7JCB2 W8B7U6 A0A0C9PJT1 K7JB64 A0A0J7K3V6 A0A087SUZ9 A0A087T3I4 A0A182H681 A0A1Z5KVF1 Q9XZR7 A1BPH1 A0A0J7K3C0 A0A0S7KB76 A0A087T293 A0A0J7K313 A0A0K3CWF3 A0A3B3IK80 A0A1W7R6D0 A0A0S7KAF1 L7MMB3 A0A0P6ATU6 A0A0J7KTY6 A0A1B6JW27 A0A147BST7 A0A1B6E3H6 A0A1L8DJ04 W4Y9K9 K7JCB3 L7LU37 A0A2G8JTJ5 A0A3B1JJM2 A0A2G8KXK5
Pubmed
EMBL
GEZM01035170
JAV83377.1
GALX01000704
JAB67762.1
GDHC01008563
JAQ10066.1
+ More
GEZM01035168 JAV83378.1 GEZM01001047 JAV98195.1 GEZM01099475 JAV53313.1 GEZM01099943 GEZM01099942 GEZM01099938 JAV53086.1 GBHO01041895 GBHO01025146 GBHO01025145 GBHO01025144 GBHO01025143 GBHO01014600 GBHO01014569 JAG01709.1 JAG18458.1 JAG18459.1 JAG18460.1 JAG18461.1 JAG29004.1 JAG29035.1 GEBQ01007184 GEBQ01002795 JAT32793.1 JAT37182.1 LBMM01018582 KMQ83753.1 GEZM01100409 GEZM01100408 GEZM01100407 GEZM01100406 GEZM01100404 GEZM01100403 JAV52889.1 GEZM01099692 JAV53185.1 LBMM01016885 KMQ84279.1 GEZM01099479 JAV53309.1 GAMC01020690 JAB85865.1 GEBQ01014606 JAT25371.1 GEZM01046053 JAV77555.1 GEZM01019359 JAV89692.1 GFTR01008544 JAW07882.1 GBBI01004690 JAC14022.1 GFTR01008516 JAW07910.1 LBMM01010645 KMQ87338.1 GEZM01033784 GEZM01033783 GEZM01033781 GEZM01033780 GEZM01033779 GEZM01033777 GEZM01033776 GEZM01033775 GEZM01033774 GEZM01033773 GEZM01033772 JAV84145.1 GALX01005179 JAB63287.1 GFTR01008518 JAW07908.1 GFTR01008424 JAW08002.1 GEZM01073925 JAV64737.1 GBYB01006259 JAG76026.1 GECU01030157 GECU01020076 JAS77549.1 JAS87630.1 LBMM01012378 KMQ86252.1 GBHO01008766 JAG34838.1 KQ971350 KYB26855.1 LBMM01010852 KMQ87188.1 LBMM01010896 KMQ87169.1 GBBI01004637 JAC14075.1 GEZM01005662 GEZM01005661 JAV96000.1 LBMM01011454 KMQ86828.1 LBMM01004568 KMQ92283.1 GFTR01008089 JAW08337.1 JXUM01014148 KQ560395 KXJ82633.1 LBMM01009042 KMQ88444.1 LBMM01009043 KMQ88443.1 KQ972957 KXZ75714.1 GALX01005355 JAB63111.1 GEZM01083034 JAV61205.1 GEZM01020516 GEZM01020515 JAV89256.1 LBMM01011641 KMQ86727.1 GFTR01008489 JAW07937.1 LBMM01013676 KMQ85528.1 GEZM01018686 JAV90062.1 GEZM01018688 GEZM01018687 GEZM01018685 GEZM01018684 GEZM01018683 GEZM01018682 JAV90067.1 GBHO01002002 GBHO01001990 JAG41602.1 JAG41614.1 GBHO01042757 JAG00847.1 LBMM01014647 KMQ85099.1 GBHO01043356 JAG00248.1 GDHC01020090 JAP98538.1 LBMM01017485 KMQ84071.1 GDHC01010105 JAQ08524.1 GECL01003261 JAP02863.1 LBMM01014329 KMQ85218.1 GEZM01046161 JAV77473.1 GDHC01007507 JAQ11122.1 GFTR01008241 JAW08185.1 KQ972733 KXZ75794.1 CH892681 KRK05548.1 LBMM01017182 KMQ84177.1 GBBI01004634 JAC14078.1 AAZX01023814 GAMC01011885 GAMC01011883 JAB94672.1 GBYB01001213 JAG70980.1 AAZX01004227 LBMM01014677 KMQ85083.1 KK112080 KFM56688.1 KK113242 KFM59673.1 JXUM01028079 KQ560809 KXJ80804.1 GFJQ02008169 JAV98800.1 AJ133521 CAB39733.1 DQ408427 ABD72267.1 LBMM01015374 KMQ84832.1 GBYX01214978 JAO74495.1 KK113062 KFM59232.1 LBMM01015404 KMQ84823.1 LN877232 CTR11712.1 GEHC01000926 JAV46719.1 GBYX01214976 JAO74497.1 GACK01000610 JAA64424.1 GDIP01038436 JAM65279.1 LBMM01003245 KMQ93756.1 GECU01004320 JAT03387.1 GEGO01001548 JAR93856.1 GEDC01004851 JAS32447.1 GFDF01007737 JAV06347.1 AAGJ04130933 AAZX01023348 GACK01009937 JAA55097.1 MRZV01001284 PIK39040.1 MRZV01000315 PIK52746.1
GEZM01035168 JAV83378.1 GEZM01001047 JAV98195.1 GEZM01099475 JAV53313.1 GEZM01099943 GEZM01099942 GEZM01099938 JAV53086.1 GBHO01041895 GBHO01025146 GBHO01025145 GBHO01025144 GBHO01025143 GBHO01014600 GBHO01014569 JAG01709.1 JAG18458.1 JAG18459.1 JAG18460.1 JAG18461.1 JAG29004.1 JAG29035.1 GEBQ01007184 GEBQ01002795 JAT32793.1 JAT37182.1 LBMM01018582 KMQ83753.1 GEZM01100409 GEZM01100408 GEZM01100407 GEZM01100406 GEZM01100404 GEZM01100403 JAV52889.1 GEZM01099692 JAV53185.1 LBMM01016885 KMQ84279.1 GEZM01099479 JAV53309.1 GAMC01020690 JAB85865.1 GEBQ01014606 JAT25371.1 GEZM01046053 JAV77555.1 GEZM01019359 JAV89692.1 GFTR01008544 JAW07882.1 GBBI01004690 JAC14022.1 GFTR01008516 JAW07910.1 LBMM01010645 KMQ87338.1 GEZM01033784 GEZM01033783 GEZM01033781 GEZM01033780 GEZM01033779 GEZM01033777 GEZM01033776 GEZM01033775 GEZM01033774 GEZM01033773 GEZM01033772 JAV84145.1 GALX01005179 JAB63287.1 GFTR01008518 JAW07908.1 GFTR01008424 JAW08002.1 GEZM01073925 JAV64737.1 GBYB01006259 JAG76026.1 GECU01030157 GECU01020076 JAS77549.1 JAS87630.1 LBMM01012378 KMQ86252.1 GBHO01008766 JAG34838.1 KQ971350 KYB26855.1 LBMM01010852 KMQ87188.1 LBMM01010896 KMQ87169.1 GBBI01004637 JAC14075.1 GEZM01005662 GEZM01005661 JAV96000.1 LBMM01011454 KMQ86828.1 LBMM01004568 KMQ92283.1 GFTR01008089 JAW08337.1 JXUM01014148 KQ560395 KXJ82633.1 LBMM01009042 KMQ88444.1 LBMM01009043 KMQ88443.1 KQ972957 KXZ75714.1 GALX01005355 JAB63111.1 GEZM01083034 JAV61205.1 GEZM01020516 GEZM01020515 JAV89256.1 LBMM01011641 KMQ86727.1 GFTR01008489 JAW07937.1 LBMM01013676 KMQ85528.1 GEZM01018686 JAV90062.1 GEZM01018688 GEZM01018687 GEZM01018685 GEZM01018684 GEZM01018683 GEZM01018682 JAV90067.1 GBHO01002002 GBHO01001990 JAG41602.1 JAG41614.1 GBHO01042757 JAG00847.1 LBMM01014647 KMQ85099.1 GBHO01043356 JAG00248.1 GDHC01020090 JAP98538.1 LBMM01017485 KMQ84071.1 GDHC01010105 JAQ08524.1 GECL01003261 JAP02863.1 LBMM01014329 KMQ85218.1 GEZM01046161 JAV77473.1 GDHC01007507 JAQ11122.1 GFTR01008241 JAW08185.1 KQ972733 KXZ75794.1 CH892681 KRK05548.1 LBMM01017182 KMQ84177.1 GBBI01004634 JAC14078.1 AAZX01023814 GAMC01011885 GAMC01011883 JAB94672.1 GBYB01001213 JAG70980.1 AAZX01004227 LBMM01014677 KMQ85083.1 KK112080 KFM56688.1 KK113242 KFM59673.1 JXUM01028079 KQ560809 KXJ80804.1 GFJQ02008169 JAV98800.1 AJ133521 CAB39733.1 DQ408427 ABD72267.1 LBMM01015374 KMQ84832.1 GBYX01214978 JAO74495.1 KK113062 KFM59232.1 LBMM01015404 KMQ84823.1 LN877232 CTR11712.1 GEHC01000926 JAV46719.1 GBYX01214976 JAO74497.1 GACK01000610 JAA64424.1 GDIP01038436 JAM65279.1 LBMM01003245 KMQ93756.1 GECU01004320 JAT03387.1 GEGO01001548 JAR93856.1 GEDC01004851 JAS32447.1 GFDF01007737 JAV06347.1 AAGJ04130933 AAZX01023348 GACK01009937 JAA55097.1 MRZV01001284 PIK39040.1 MRZV01000315 PIK52746.1
Proteomes
Pfam
Interpro
IPR041373
RT_RNaseH
+ More
IPR000477 RT_dom
IPR001969 Aspartic_peptidase_AS
IPR021109 Peptidase_aspartic_dom_sf
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR034122 Retropepsin-like_bacterial
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR018061 Retropepsins
IPR005162 Retrotrans_gag_dom
IPR003034 SAP_dom
IPR021896 Transposase_37
IPR019103 Peptidase_aspartic_DDI1-type
IPR036361 SAP_dom_sf
IPR002492 Transposase_Tc1-like
IPR009057 Homeobox-like_sf
IPR038717 Tc1-like_DDE_dom
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR003309 SCAN_dom
IPR019787 Znf_PHD-finger
IPR000477 RT_dom
IPR001969 Aspartic_peptidase_AS
IPR021109 Peptidase_aspartic_dom_sf
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR034122 Retropepsin-like_bacterial
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR018061 Retropepsins
IPR005162 Retrotrans_gag_dom
IPR003034 SAP_dom
IPR021896 Transposase_37
IPR019103 Peptidase_aspartic_DDI1-type
IPR036361 SAP_dom_sf
IPR002492 Transposase_Tc1-like
IPR009057 Homeobox-like_sf
IPR038717 Tc1-like_DDE_dom
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR003309 SCAN_dom
IPR019787 Znf_PHD-finger
SUPFAM
Gene 3D
ProteinModelPortal
A0A1Y1MJD1
V5H0H7
A0A146LTK8
A0A1Y1MED9
A0A1Y1NMV4
A0A1Y1JW20
+ More
A0A1Y1K180 A0A0A9XMW3 A0A1B6MMN8 A0A0J7JZY0 A0A1Y1JUB3 A0A1Y1K0I6 A0A0J7K1V9 A0A1Y1JVH7 W8AD09 A0A1B6LNQ5 A0A1Y1LZS5 A0A1Y1MVI4 A0A224XH29 A0A023EY78 A0A224XHI1 A0A0J7KA76 A0A1Y1MIJ6 V5GNV6 A0A224XDN8 A0A224XHE2 A0A1Y1L0V4 A0A0C9QRC9 A0A1B6IL28 A0A0J7K794 A0A0A9YUQ6 A0A139WG74 A0A0J7K9T8 A0A0J7KA70 A0A023EXS9 A0A1Y1NGJ5 A0A0J7K8R1 A0A0J7KPJ7 A0A224X7A2 A0A182GM87 A0A0J7KE06 A0A0J7KDB6 A0A139W8U5 V5I839 A0A1Y1KNF8 A0A1Y1MZ56 A0A0J7K8Q7 A0A224X632 A0A0J7K4Z4 A0A1Y1MWM5 A0A1Y1N3D6 A0A0A9Z8Q1 A0A0A9VXA9 A0A0J7K489 A0A0A9W1A6 A0A146KR86 A0A0J7K1N1 A0A146LKA7 A0A0V0G672 A0A0J7K441 A0A1Y1LV33 A0A146LWQ9 A0A224X6V5 A0A139W930 A0A0R1E8A2 A0A0J7K1I1 A0A023EY81 K7JCB2 W8B7U6 A0A0C9PJT1 K7JB64 A0A0J7K3V6 A0A087SUZ9 A0A087T3I4 A0A182H681 A0A1Z5KVF1 Q9XZR7 A1BPH1 A0A0J7K3C0 A0A0S7KB76 A0A087T293 A0A0J7K313 A0A0K3CWF3 A0A3B3IK80 A0A1W7R6D0 A0A0S7KAF1 L7MMB3 A0A0P6ATU6 A0A0J7KTY6 A0A1B6JW27 A0A147BST7 A0A1B6E3H6 A0A1L8DJ04 W4Y9K9 K7JCB3 L7LU37 A0A2G8JTJ5 A0A3B1JJM2 A0A2G8KXK5
A0A1Y1K180 A0A0A9XMW3 A0A1B6MMN8 A0A0J7JZY0 A0A1Y1JUB3 A0A1Y1K0I6 A0A0J7K1V9 A0A1Y1JVH7 W8AD09 A0A1B6LNQ5 A0A1Y1LZS5 A0A1Y1MVI4 A0A224XH29 A0A023EY78 A0A224XHI1 A0A0J7KA76 A0A1Y1MIJ6 V5GNV6 A0A224XDN8 A0A224XHE2 A0A1Y1L0V4 A0A0C9QRC9 A0A1B6IL28 A0A0J7K794 A0A0A9YUQ6 A0A139WG74 A0A0J7K9T8 A0A0J7KA70 A0A023EXS9 A0A1Y1NGJ5 A0A0J7K8R1 A0A0J7KPJ7 A0A224X7A2 A0A182GM87 A0A0J7KE06 A0A0J7KDB6 A0A139W8U5 V5I839 A0A1Y1KNF8 A0A1Y1MZ56 A0A0J7K8Q7 A0A224X632 A0A0J7K4Z4 A0A1Y1MWM5 A0A1Y1N3D6 A0A0A9Z8Q1 A0A0A9VXA9 A0A0J7K489 A0A0A9W1A6 A0A146KR86 A0A0J7K1N1 A0A146LKA7 A0A0V0G672 A0A0J7K441 A0A1Y1LV33 A0A146LWQ9 A0A224X6V5 A0A139W930 A0A0R1E8A2 A0A0J7K1I1 A0A023EY81 K7JCB2 W8B7U6 A0A0C9PJT1 K7JB64 A0A0J7K3V6 A0A087SUZ9 A0A087T3I4 A0A182H681 A0A1Z5KVF1 Q9XZR7 A1BPH1 A0A0J7K3C0 A0A0S7KB76 A0A087T293 A0A0J7K313 A0A0K3CWF3 A0A3B3IK80 A0A1W7R6D0 A0A0S7KAF1 L7MMB3 A0A0P6ATU6 A0A0J7KTY6 A0A1B6JW27 A0A147BST7 A0A1B6E3H6 A0A1L8DJ04 W4Y9K9 K7JCB3 L7LU37 A0A2G8JTJ5 A0A3B1JJM2 A0A2G8KXK5
PDB
4OL8
E-value=1.48366e-70,
Score=677
Ontologies
GO
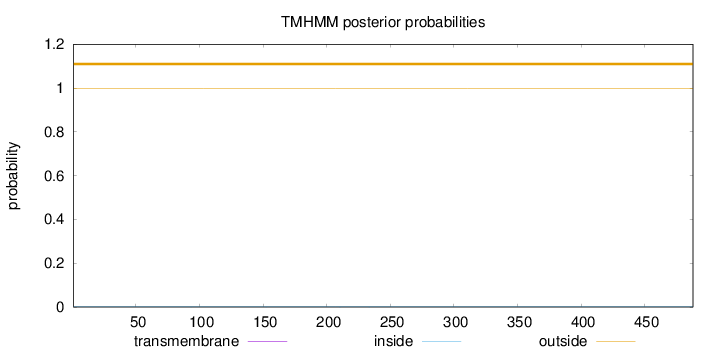
Topology
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00063
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00080
outside
1 - 488
Population Genetic Test Statistics
Pi
13.553717
Theta
20.650132
Tajima's D
-1.387926
CLR
140.123041
CSRT
0.072196390180491
Interpretation
Uncertain
