Gene
KWMTBOMO03123
Pre Gene Modal
BGIBMGA005791
Annotation
PREDICTED:_adenosine_monophosphate-protein_transferase_FICD_homolog_[Bombyx_mori]
Full name
Protein adenylyltransferase Fic
Alternative Name
De-AMPylase Fic
Location in the cell
Mitochondrial Reliability : 1.487 Nuclear Reliability : 1.401
Sequence
CDS
ATGTTGTACTTAGGTAAAACGAGCGAAGGCGAAAAAATGAAGTGTTCGTGCAAAAATGAAATATTCACCAAAATGATAGTGGATAAGTGCAGGACCGTAGTCATACTCTCTAGTATGGTGTGTACACTAGCTGTTGTTATATCTTTACACAAATTATTAACTTATGACATTAGAATTACCAAACCATTTTCCCCTATACCAGAAGGGCTGTACGGGAACTACTTAGAACCAATTTCAGATGAGAATCCCGTACCTATAGTACCTAAGGCACAAGATAAAGCTCGGGATGCGGAAGCTGTGGTGTCTTTAAATGCTGCATTAGAAATGAAGAAACATGGAAAGTCCGACAAAGCATTAAAATTATTTCAACATGCATTCGCTCTGTCACCAAAACATGCAGATATTTTAAATCACTATGGAGAGTTCTTAGAAGATACTAAAAAAGATATAGTAAAAGCAGACCAGCTTTATACATTAGCTTTAACTAATTACCCTGATCACACTGGTGCTTTAACGAACAGACAAAGAACAGCCAGCATTGTTGAAAACTTAGACAGAGAAATGTTGAGGAAAATTGATGATAAAAGAGATGCACTATCTTCAATACCAGAAAACAATTCAGCATTACGAAGAGCAAAAAAGGAGGCATATTTCCAACATATTTATCACACAGTAGCCATAGAAGGTAATACAATGACATTGCAACAAACAAGAAGTATTCTAGAGACTCGGATTGCTATATCGGGCAAAAGCATAGATGAGCACAATGAAATTCTAGGCTTAGATGCAGCAATGAAGTACATAAATTCTACACTTCTGTACAGATTACGAGACATAACCATGGGTGATATTCTTGAGATTCATAAGAGGGTGTTAGGGCATGTAGACCCAGTGGAAGGAGGGAACTTCCGCAGAACACAGGTGTATGTTGGTGGTCACATTCCACCTGGCCCCTTAGAGATCCAAGGCCTAATGATACAGTTTATGGAATGGCTTAACTCTGAAGATGCTTTGGATTTACATCCAGTAAGGTACGCAGCCCTGGCGCACTACAAACTAGTGCACATCCACCCGTTCGTGGACGGCAACGGACGCACGTCGCGGCTCCTCATGAACCTGCTGCTGATGCAGGCCGGATACCCCCCCGTCATCATCGCGAAACAACACCGCCACCTTTACTACCAGCATTTGCAGACCGCTAACGAGGGAGATGTTAGACCTTTTGTTAGGTTTATAGCACAATCAACAGAGCGAACTCTAAACCTGTACCTGTGGGCGACCAGCGAGTTCTCCCACACGGTCCCGGCCATCGGCAAACCTCATATACTTACCGAAGATGAATTCGACGATACAATTCTCTGA
Protein
MLYLGKTSEGEKMKCSCKNEIFTKMIVDKCRTVVILSSMVCTLAVVISLHKLLTYDIRITKPFSPIPEGLYGNYLEPISDENPVPIVPKAQDKARDAEAVVSLNAALEMKKHGKSDKALKLFQHAFALSPKHADILNHYGEFLEDTKKDIVKADQLYTLALTNYPDHTGALTNRQRTASIVENLDREMLRKIDDKRDALSSIPENNSALRRAKKEAYFQHIYHTVAIEGNTMTLQQTRSILETRIAISGKSIDEHNEILGLDAAMKYINSTLLYRLRDITMGDILEIHKRVLGHVDPVEGGNFRRTQVYVGGHIPPGPLEIQGLMIQFMEWLNSEDALDLHPVRYAALAHYKLVHIHPFVDGNGRTSRLLMNLLLMQAGYPPVIIAKQHRHLYYQHLQTANEGDVRPFVRFIAQSTERTLNLYLWATSEFSHTVPAIGKPHILTEDEFDDTIL
Summary
Description
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-255 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-251 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-250 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-236 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-247 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (PubMed:29089387). The side chain of Glu-247 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP (PubMed:25395623, PubMed:29089387). In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it (PubMed:29089387). In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (PubMed:29089387).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-261 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-252 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-251 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-250 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-236 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-247 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (PubMed:29089387). The side chain of Glu-247 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP (PubMed:25395623, PubMed:29089387). In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it (PubMed:29089387). In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (PubMed:29089387).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-261 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Protein that can both mediate the addition of adenosine 5'-monophosphate (AMP) to specific residues of target proteins (AMPylation), and the removal of the same modification from target proteins (de-AMPylation), depending on the context (By similarity). The side chain of Glu-252 determines which of the two opposing activities (AMPylase or de-AMPylase) will take place (By similarity). Acts as a key regulator of the unfolded protein response (UPR) by mediating AMPylation or de-AMPylation of Hsc70-3/BiP. In unstressed cells, acts as an adenylyltransferase by mediating AMPylation of Hsc70-3/BiP at 'Thr-518', thereby inactivating it. In response to endoplasmic reticulum stress, acts as a phosphodiesterase by mediating removal of ATP (de-AMPylation) from Hsc70-3/BiP at 'Thr-518', leading to restore HSPA5/BiP activity (By similarity).
Catalytic Activity
ATP + L-tyrosyl-[protein] = diphosphate + O-(5'-adenylyl)-L-tyrosyl-[protein]
ATP + L-threonyl-[protein] = 3-O-(5'-adenylyl)-L-threonyl-[protein] + diphosphate
3-O-(5'-adenylyl)-L-threonyl-[protein] + H2O = AMP + H(+) + L-threonyl-[protein]
ATP + L-threonyl-[protein] = 3-O-(5'-adenylyl)-L-threonyl-[protein] + diphosphate
3-O-(5'-adenylyl)-L-threonyl-[protein] + H2O = AMP + H(+) + L-threonyl-[protein]
Subunit
Homodimer.
Homodimer; homodimerization may regulate adenylyltransferase and phosphodiesterase activities.
Homodimer; homodimerization may regulate adenylyltransferase and phosphodiesterase activities.
Similarity
Belongs to the fic family.
Keywords
ATP-binding
Complete proteome
Hydrolase
Membrane
Nucleotide-binding
Nucleotidyltransferase
Reference proteome
Repeat
TPR repeat
Transferase
Transmembrane
Transmembrane helix
Feature
chain Protein adenylyltransferase Fic
Uniprot
A0A2H1V6C4
A0A0L7LNN7
A0A2W1BIA5
A0A2A4J4N4
H9J8E9
A0A194PP13
+ More
A0A0N0PBC2 A0A212EJ58 A0A0N0BJ80 A0A310SEF7 A0A2J7R3H1 A0A088A303 E9IB73 A0A2A3EA94 A0A0L7QXJ4 A0A195B6Z9 E2AZD6 A0A195CPH8 F4WG96 A0A158NI65 A0A151JX74 A0A026WTM1 A0A154P523 A0A195EGM3 E2C6W3 A0A067QSR1 A0A151WJE0 A0A182Y5M9 A0A1Q3F7G6 Q7Q1Y3 Q17A75 B0W429 A0A182PB18 A0A182RRG4 A0A1Q3F812 A0A1B6DIV6 A0A2M3Z7N7 A0A182IDX8 A0A182WHK6 A0A182UG26 A0A182LV45 A0A182K1B5 A0A182UYL7 A0A182NUZ1 A0A182XCJ8 A0A182F232 A0A2M3Z6Q4 A0A2M3Z6U7 A0A1A9XTP6 A0A1B0C7A9 A0A182Q5G9 A0A2M4BJ19 A0A2M4BJ35 A0A2M4BJ85 A0A084VDU0 A0A1B0FEY9 A0A182IQP3 A0A1A9X085 A0A1B6ESG1 A0A1A9V7F5 A0A1B6L271 A0A1A9Z5R5 A0A0L0BRZ8 E0VQJ4 A0A0K8TS66 A0A336KT82 A0A0K8UK48 A0A034VRR1 U5EIG0 A0A336MBF3 W8BKS6 B4P0E1 B3N5J3 A0A0A9WH54 B4JBN5 B4LQT7 B4KFW6 A0A3B0KD51 A0A0J9QXP8 B4I1V5 B4MUQ2 B4Q4M7 Q8SWV6 A0A0P4VN55 A0A1I8MML5 Q29JP8 B4GJC1 A0A1W4V8P6 A0A1I8NSW1 A0A1J1HT59 W5JQ21 B3MK83 A0A0P5K1N6 A0A0P4YAA9 A0A0P5GMF2 A0A165AFP9 A0A0P6BEX1 A0A0V0G742 A0A0M5IXU3 A0A0M4E0A6
A0A0N0PBC2 A0A212EJ58 A0A0N0BJ80 A0A310SEF7 A0A2J7R3H1 A0A088A303 E9IB73 A0A2A3EA94 A0A0L7QXJ4 A0A195B6Z9 E2AZD6 A0A195CPH8 F4WG96 A0A158NI65 A0A151JX74 A0A026WTM1 A0A154P523 A0A195EGM3 E2C6W3 A0A067QSR1 A0A151WJE0 A0A182Y5M9 A0A1Q3F7G6 Q7Q1Y3 Q17A75 B0W429 A0A182PB18 A0A182RRG4 A0A1Q3F812 A0A1B6DIV6 A0A2M3Z7N7 A0A182IDX8 A0A182WHK6 A0A182UG26 A0A182LV45 A0A182K1B5 A0A182UYL7 A0A182NUZ1 A0A182XCJ8 A0A182F232 A0A2M3Z6Q4 A0A2M3Z6U7 A0A1A9XTP6 A0A1B0C7A9 A0A182Q5G9 A0A2M4BJ19 A0A2M4BJ35 A0A2M4BJ85 A0A084VDU0 A0A1B0FEY9 A0A182IQP3 A0A1A9X085 A0A1B6ESG1 A0A1A9V7F5 A0A1B6L271 A0A1A9Z5R5 A0A0L0BRZ8 E0VQJ4 A0A0K8TS66 A0A336KT82 A0A0K8UK48 A0A034VRR1 U5EIG0 A0A336MBF3 W8BKS6 B4P0E1 B3N5J3 A0A0A9WH54 B4JBN5 B4LQT7 B4KFW6 A0A3B0KD51 A0A0J9QXP8 B4I1V5 B4MUQ2 B4Q4M7 Q8SWV6 A0A0P4VN55 A0A1I8MML5 Q29JP8 B4GJC1 A0A1W4V8P6 A0A1I8NSW1 A0A1J1HT59 W5JQ21 B3MK83 A0A0P5K1N6 A0A0P4YAA9 A0A0P5GMF2 A0A165AFP9 A0A0P6BEX1 A0A0V0G742 A0A0M5IXU3 A0A0M4E0A6
EC Number
2.7.7.n1
Pubmed
26227816
28756777
19121390
26354079
22118469
21282665
+ More
20798317 21719571 21347285 24508170 30249741 24845553 25244985 12364791 17510324 24438588 26108605 20566863 26369729 25348373 24495485 17994087 25401762 22936249 10731132 12537572 12537569 19503829 25395623 29089387 27129103 25315136 15632085 20920257 23761445
20798317 21719571 21347285 24508170 30249741 24845553 25244985 12364791 17510324 24438588 26108605 20566863 26369729 25348373 24495485 17994087 25401762 22936249 10731132 12537572 12537569 19503829 25395623 29089387 27129103 25315136 15632085 20920257 23761445
EMBL
ODYU01000919
SOQ36405.1
JTDY01000463
KOB77062.1
KZ150027
PZC74782.1
+ More
NWSH01003247 PCG66696.1 BABH01027325 KQ459597 KPI95171.1 KQ460970 KPJ10185.1 AGBW02014532 OWR41500.1 KQ435719 KOX78804.1 KQ770178 OAD52675.1 NEVH01007819 PNF35388.1 GL762111 EFZ22212.1 KZ288322 PBC28212.1 KQ414704 KOC63330.1 KQ976579 KYM80035.1 GL444207 EFN61175.1 KQ977481 KYN02397.1 GL888128 EGI66863.1 ADTU01016347 KQ981578 KYN39744.1 KK107111 QOIP01000011 EZA59011.1 RLU17244.1 KQ434809 KZC06424.1 KQ978957 KYN27047.1 GL453216 EFN76317.1 KK869029 KDR12881.1 KQ983039 KYQ47972.1 GFDL01011539 JAV23506.1 AAAB01008980 EAA13788.4 CH477339 DS231834 GFDL01011402 JAV23643.1 GEDC01011711 JAS25587.1 GGFM01003795 MBW24546.1 APCN01004537 AXCM01001697 GGFM01003434 MBW24185.1 GGFM01003473 MBW24224.1 JXJN01027972 AXCN02000753 GGFJ01003925 MBW53066.1 GGFJ01003924 MBW53065.1 GGFJ01003923 MBW53064.1 ATLV01011788 KE524721 KFB36134.1 CCAG010008989 GECZ01028887 JAS40882.1 GEBQ01022343 JAT17634.1 JRES01001455 KNC22773.1 DS235430 EEB15650.1 GDAI01000612 JAI16991.1 UFQS01000861 UFQT01000861 SSX07342.1 SSX27684.1 GDHF01025393 JAI26921.1 GAKP01014165 GAKP01014162 GAKP01014160 JAC44792.1 GANO01002662 JAB57209.1 SSX07340.1 SSX27682.1 GAMC01016391 JAB90164.1 CM000157 CH954177 GBHO01035827 GBRD01006244 JAG07777.1 JAG59577.1 CH916368 CH940649 CH933807 OUUW01000010 SPP86170.1 CM002910 KMY88514.1 CH480820 CH963857 CM000361 AE014134 AY095059 GDKW01002277 JAI54318.1 CH379062 CH479184 CVRI01000020 CRK90564.1 ADMH02000487 ETN66236.1 CH902620 GDIQ01191008 JAK60717.1 GDIP01231280 JAI92121.1 GDIQ01239654 GDIQ01036861 JAK12071.1 LRGB01000626 KZS17582.1 GDIP01030345 JAM73370.1 GECL01002596 JAP03528.1 CP012523 ALC37998.1 ALC38491.1
NWSH01003247 PCG66696.1 BABH01027325 KQ459597 KPI95171.1 KQ460970 KPJ10185.1 AGBW02014532 OWR41500.1 KQ435719 KOX78804.1 KQ770178 OAD52675.1 NEVH01007819 PNF35388.1 GL762111 EFZ22212.1 KZ288322 PBC28212.1 KQ414704 KOC63330.1 KQ976579 KYM80035.1 GL444207 EFN61175.1 KQ977481 KYN02397.1 GL888128 EGI66863.1 ADTU01016347 KQ981578 KYN39744.1 KK107111 QOIP01000011 EZA59011.1 RLU17244.1 KQ434809 KZC06424.1 KQ978957 KYN27047.1 GL453216 EFN76317.1 KK869029 KDR12881.1 KQ983039 KYQ47972.1 GFDL01011539 JAV23506.1 AAAB01008980 EAA13788.4 CH477339 DS231834 GFDL01011402 JAV23643.1 GEDC01011711 JAS25587.1 GGFM01003795 MBW24546.1 APCN01004537 AXCM01001697 GGFM01003434 MBW24185.1 GGFM01003473 MBW24224.1 JXJN01027972 AXCN02000753 GGFJ01003925 MBW53066.1 GGFJ01003924 MBW53065.1 GGFJ01003923 MBW53064.1 ATLV01011788 KE524721 KFB36134.1 CCAG010008989 GECZ01028887 JAS40882.1 GEBQ01022343 JAT17634.1 JRES01001455 KNC22773.1 DS235430 EEB15650.1 GDAI01000612 JAI16991.1 UFQS01000861 UFQT01000861 SSX07342.1 SSX27684.1 GDHF01025393 JAI26921.1 GAKP01014165 GAKP01014162 GAKP01014160 JAC44792.1 GANO01002662 JAB57209.1 SSX07340.1 SSX27682.1 GAMC01016391 JAB90164.1 CM000157 CH954177 GBHO01035827 GBRD01006244 JAG07777.1 JAG59577.1 CH916368 CH940649 CH933807 OUUW01000010 SPP86170.1 CM002910 KMY88514.1 CH480820 CH963857 CM000361 AE014134 AY095059 GDKW01002277 JAI54318.1 CH379062 CH479184 CVRI01000020 CRK90564.1 ADMH02000487 ETN66236.1 CH902620 GDIQ01191008 JAK60717.1 GDIP01231280 JAI92121.1 GDIQ01239654 GDIQ01036861 JAK12071.1 LRGB01000626 KZS17582.1 GDIP01030345 JAM73370.1 GECL01002596 JAP03528.1 CP012523 ALC37998.1 ALC38491.1
Proteomes
UP000037510
UP000218220
UP000005204
UP000053268
UP000053240
UP000007151
+ More
UP000053105 UP000235965 UP000005203 UP000242457 UP000053825 UP000078540 UP000000311 UP000078542 UP000007755 UP000005205 UP000078541 UP000053097 UP000279307 UP000076502 UP000078492 UP000008237 UP000027135 UP000075809 UP000076408 UP000007062 UP000008820 UP000002320 UP000075885 UP000075900 UP000075840 UP000075920 UP000075902 UP000075883 UP000075881 UP000075903 UP000075884 UP000076407 UP000069272 UP000092443 UP000092460 UP000075886 UP000030765 UP000092444 UP000075880 UP000091820 UP000078200 UP000092445 UP000037069 UP000009046 UP000002282 UP000008711 UP000001070 UP000008792 UP000009192 UP000268350 UP000001292 UP000007798 UP000000304 UP000000803 UP000095301 UP000001819 UP000008744 UP000192221 UP000095300 UP000183832 UP000000673 UP000007801 UP000076858 UP000092553
UP000053105 UP000235965 UP000005203 UP000242457 UP000053825 UP000078540 UP000000311 UP000078542 UP000007755 UP000005205 UP000078541 UP000053097 UP000279307 UP000076502 UP000078492 UP000008237 UP000027135 UP000075809 UP000076408 UP000007062 UP000008820 UP000002320 UP000075885 UP000075900 UP000075840 UP000075920 UP000075902 UP000075883 UP000075881 UP000075903 UP000075884 UP000076407 UP000069272 UP000092443 UP000092460 UP000075886 UP000030765 UP000092444 UP000075880 UP000091820 UP000078200 UP000092445 UP000037069 UP000009046 UP000002282 UP000008711 UP000001070 UP000008792 UP000009192 UP000268350 UP000001292 UP000007798 UP000000304 UP000000803 UP000095301 UP000001819 UP000008744 UP000192221 UP000095300 UP000183832 UP000000673 UP000007801 UP000076858 UP000092553
Interpro
Gene 3D
ProteinModelPortal
A0A2H1V6C4
A0A0L7LNN7
A0A2W1BIA5
A0A2A4J4N4
H9J8E9
A0A194PP13
+ More
A0A0N0PBC2 A0A212EJ58 A0A0N0BJ80 A0A310SEF7 A0A2J7R3H1 A0A088A303 E9IB73 A0A2A3EA94 A0A0L7QXJ4 A0A195B6Z9 E2AZD6 A0A195CPH8 F4WG96 A0A158NI65 A0A151JX74 A0A026WTM1 A0A154P523 A0A195EGM3 E2C6W3 A0A067QSR1 A0A151WJE0 A0A182Y5M9 A0A1Q3F7G6 Q7Q1Y3 Q17A75 B0W429 A0A182PB18 A0A182RRG4 A0A1Q3F812 A0A1B6DIV6 A0A2M3Z7N7 A0A182IDX8 A0A182WHK6 A0A182UG26 A0A182LV45 A0A182K1B5 A0A182UYL7 A0A182NUZ1 A0A182XCJ8 A0A182F232 A0A2M3Z6Q4 A0A2M3Z6U7 A0A1A9XTP6 A0A1B0C7A9 A0A182Q5G9 A0A2M4BJ19 A0A2M4BJ35 A0A2M4BJ85 A0A084VDU0 A0A1B0FEY9 A0A182IQP3 A0A1A9X085 A0A1B6ESG1 A0A1A9V7F5 A0A1B6L271 A0A1A9Z5R5 A0A0L0BRZ8 E0VQJ4 A0A0K8TS66 A0A336KT82 A0A0K8UK48 A0A034VRR1 U5EIG0 A0A336MBF3 W8BKS6 B4P0E1 B3N5J3 A0A0A9WH54 B4JBN5 B4LQT7 B4KFW6 A0A3B0KD51 A0A0J9QXP8 B4I1V5 B4MUQ2 B4Q4M7 Q8SWV6 A0A0P4VN55 A0A1I8MML5 Q29JP8 B4GJC1 A0A1W4V8P6 A0A1I8NSW1 A0A1J1HT59 W5JQ21 B3MK83 A0A0P5K1N6 A0A0P4YAA9 A0A0P5GMF2 A0A165AFP9 A0A0P6BEX1 A0A0V0G742 A0A0M5IXU3 A0A0M4E0A6
A0A0N0PBC2 A0A212EJ58 A0A0N0BJ80 A0A310SEF7 A0A2J7R3H1 A0A088A303 E9IB73 A0A2A3EA94 A0A0L7QXJ4 A0A195B6Z9 E2AZD6 A0A195CPH8 F4WG96 A0A158NI65 A0A151JX74 A0A026WTM1 A0A154P523 A0A195EGM3 E2C6W3 A0A067QSR1 A0A151WJE0 A0A182Y5M9 A0A1Q3F7G6 Q7Q1Y3 Q17A75 B0W429 A0A182PB18 A0A182RRG4 A0A1Q3F812 A0A1B6DIV6 A0A2M3Z7N7 A0A182IDX8 A0A182WHK6 A0A182UG26 A0A182LV45 A0A182K1B5 A0A182UYL7 A0A182NUZ1 A0A182XCJ8 A0A182F232 A0A2M3Z6Q4 A0A2M3Z6U7 A0A1A9XTP6 A0A1B0C7A9 A0A182Q5G9 A0A2M4BJ19 A0A2M4BJ35 A0A2M4BJ85 A0A084VDU0 A0A1B0FEY9 A0A182IQP3 A0A1A9X085 A0A1B6ESG1 A0A1A9V7F5 A0A1B6L271 A0A1A9Z5R5 A0A0L0BRZ8 E0VQJ4 A0A0K8TS66 A0A336KT82 A0A0K8UK48 A0A034VRR1 U5EIG0 A0A336MBF3 W8BKS6 B4P0E1 B3N5J3 A0A0A9WH54 B4JBN5 B4LQT7 B4KFW6 A0A3B0KD51 A0A0J9QXP8 B4I1V5 B4MUQ2 B4Q4M7 Q8SWV6 A0A0P4VN55 A0A1I8MML5 Q29JP8 B4GJC1 A0A1W4V8P6 A0A1I8NSW1 A0A1J1HT59 W5JQ21 B3MK83 A0A0P5K1N6 A0A0P4YAA9 A0A0P5GMF2 A0A165AFP9 A0A0P6BEX1 A0A0V0G742 A0A0M5IXU3 A0A0M4E0A6
PDB
4U0Z
E-value=6.70421e-114,
Score=1051
Ontologies
GO
PANTHER
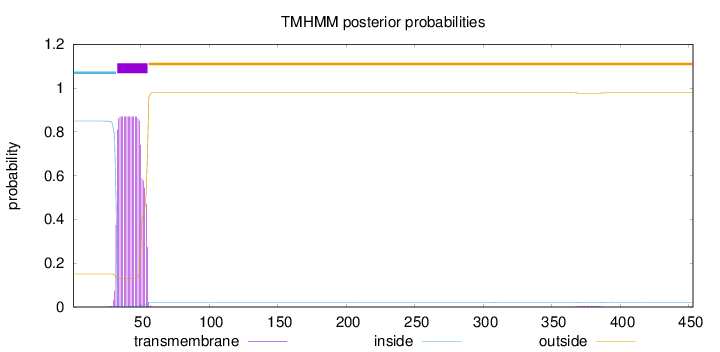
Topology
Subcellular location
Membrane
Length:
453
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.48164
Exp number, first 60 AAs:
18.40159
Total prob of N-in:
0.84899
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 453
Population Genetic Test Statistics
Pi
2.010929
Theta
6.853155
Tajima's D
-2.035326
CLR
655.388915
CSRT
0.0101994900254987
Interpretation
Possibly Positive selection
