Pre Gene Modal
BGIBMGA005777
Annotation
PREDICTED:_kinesin-like_protein_a_isoform_X2_[Bombyx_mori]
Full name
Kinesin-like protein
Location in the cell
Nuclear Reliability : 2.684
Sequence
CDS
ATGGATGACGGCACAGTTAGAATTCGCTCCGGTATCAACATCAACATTAAAAGAACAGATGGTCGGGTCCATTCTGCGATCGTGGCCTCAGTTAACTTGGAGACGCGCTCAGTTGCTGTGGAATGGTTCGAGCGCGGCGAGACCAAGGGCAAGGAGATCGAGATAGATGCCATCCTAGCGCTGAACCCTGAACTGGCGAGTGGGCCGCGGCACCACACTCCGCACCTGCAGCCGATGCCCAACAAACTAGCCAGGGATATGACGCGGCAGTCCATACCCGTTGCGACCAAAGCGCCCGTGACCAGGGTGACGCGGAACAATCAGATGCGGGTGTCGAATGTGGGCGGCGCCTACACAAACGGTCACGGCGGCGACACGACGGGGCGGCGCGGCGTCGAGAATGTACCCCCGGCACAGCTGCCGCCACAGCAGCCGCCGCCCAGTTCCAGAATCACACAGCAGTTTTCAGGTTCGGGTATTTCCAGTAGGGCGGCCGGCGTCGCGTCCACCGCGTCCGTGCCCGCCGGCGCCACCTCCGCCGTCACGTCCGCCGTCACATCCGGCGTCACGTCCGGCGTCACGTCCGGCGCGGTGCGGCGCTCCAACGTCGTCAAGGAGGTCGAGCGGCTCAAGGAGAACAGGGAGAAGAGAAGGCAGAGGCAAGCGGAACTTAAGGAAGAAAAGGAAGCCCTCATGAACCTGGACCCTGGCAACCCGAACTGGGAGTTCCTGGCGATGATCCGGGAGTACCAGAACAGCATCGAGTTCAGGCCGCTCACCGGCACCGAGCCCGTCGAGGACCACCAGATCACGGTCTGCGTGCGGAAGAGGCCGCTCAACAAGAAGGAGCTTAATAAGAAAGAGGTGGACGTGATCAGCGTCCCCACCAAGGACCAGATGATAGTCCACGAGCCCAAGAACAAAGTGGACCTCACGAAGTATTTAGAAAATCAAAAGTTTCGATTTGACTACGCGTTCGACGACTCCTGCACTAACGAAATTGTTTACAAGTACACGGCCAAGCCGCTGGTGGCGACCATCTTCGAGGGCGGCATGGCCACCTGCTTCGCGTACGGACAGACCGGCTCCGGCAAGACGCACACCATGGGCGGAGACTTCCAGGGCAAGACCCAGGACTGCAAGAAGGGCATATACGCCATGGCGGCGCGCGACGTGTTCACCTACCTGCGCGCCCCCAAGTACAAGCCGCTCAACCTCATCGTGTCCGCCTCCTTCTTCGAGATCTACTCCGGGAAGGTGTTCGACCTGCTAGCGGACAAGGCCAAGCTGAGGGTGTTGGAGGACGGCAAGCAGCAAGTGCAGATCGTGGGCCTCACCGAGAAGGTGGTGGACAACGTCGACGAGGTGCTCAAGCTCATCCAGCACGGCAACGCCGCCCGGACCTCCGGCCAGACGTCGGCCAACTCCAACTCGTCGCGCTCGCACGCCGTGTTCCAGATCGTGGTGCGGTCCCCGGGCATGCACCGCGTGCACGGGAAGTTCTCGCTCATCGACCTGGCGGGCAACGAGCGCGGCGCCGACACCTCCTCCGCCAACCGACAGACCAGAATGGAAAGCGCGGAGATCAACAAGTCCCTCCTGGCACTGAAGGAGTGCATCCGCGCGCTGGGCATGAAGGGCAACAACCACCTGCCCTTCCGCGTGTCCAAGCTCACGCAGGTCCTGCGAGACAGCTTCATCGGGGACAAGTCTCGCACCTGCATGATCGCCATGATCTCGCCGGCCATGGCCTCCTGCGAGCACAGCCTCAACACGCTCCGATACGCCGACAGGGTGAAGGAGCTCGGCACCATGGACCCGTCCCGGCGCGGCGAGTCTCCGCCGCCCGACGTGGACATGCCGCCCGCGCGGGACGACCTGGCGCACCTGCGCTCCCTCAACGAGGGCGACATGTCGGCCGAGATGTACACCTTCCACGAAGCCATCGACGAGCTGCAGCGCGCGGAGGAGGAGGTCCTGGACAACCACAAGGCCGTGTCCGACTACATGCACCACGCGCTGCTCCGCGCCAACCATCTGCTCGCCATCACCAGGGACGTGGACTACGACCAGGACGCGTACGCGACGCAGTGGGAGGAGCTGCTGAACGAGCAGCTGGCCGTGCTGGCGCAGTCCCGCGACCTGGTGGCAGAGTTCCGCACCAAGCTCACGCGCGAGGAGCACATCTCGCGCCGCATACACAACTAG
Protein
MDDGTVRIRSGININIKRTDGRVHSAIVASVNLETRSVAVEWFERGETKGKEIEIDAILALNPELASGPRHHTPHLQPMPNKLARDMTRQSIPVATKAPVTRVTRNNQMRVSNVGGAYTNGHGGDTTGRRGVENVPPAQLPPQQPPPSSRITQQFSGSGISSRAAGVASTASVPAGATSAVTSAVTSGVTSGVTSGAVRRSNVVKEVERLKENREKRRQRQAELKEEKEALMNLDPGNPNWEFLAMIREYQNSIEFRPLTGTEPVEDHQITVCVRKRPLNKKELNKKEVDVISVPTKDQMIVHEPKNKVDLTKYLENQKFRFDYAFDDSCTNEIVYKYTAKPLVATIFEGGMATCFAYGQTGSGKTHTMGGDFQGKTQDCKKGIYAMAARDVFTYLRAPKYKPLNLIVSASFFEIYSGKVFDLLADKAKLRVLEDGKQQVQIVGLTEKVVDNVDEVLKLIQHGNAARTSGQTSANSNSSRSHAVFQIVVRSPGMHRVHGKFSLIDLAGNERGADTSSANRQTRMESAEINKSLLALKECIRALGMKGNNHLPFRVSKLTQVLRDSFIGDKSRTCMIAMISPAMASCEHSLNTLRYADRVKELGTMDPSRRGESPPPDVDMPPARDDLAHLRSLNEGDMSAEMYTFHEAIDELQRAEEEVLDNHKAVSDYMHHALLRANHLLAITRDVDYDQDAYATQWEELLNEQLAVLAQSRDLVAEFRTKLTREEHISRRIHN
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Feature
chain Kinesin-like protein
Uniprot
L8B3Q2
A0A212EJ27
A1E131
H9J8D5
A0A2H1V6K0
S4PY99
+ More
A0A194PVL5 A0A154P3L6 A0A1Q3G2E6 A0A026WSQ2 A0A336K0Z8 A0A2J7RKI6 A0A1I8P567 A0A0P6GNC7 A0A1B6KR21 A0A1B6LEG6 A0A1B6D8W4 A0A336LQ14 A0A1J1HTC2 A0A1Q3G2H2 A0A158NI67 A0A336K0W4 E0VQJ3 U5ELQ3 E9IB70 A0A1Q3G213 A0A2H8TQM6 A0A1B6DU41 J9JUC2 A0A2S2PLX8 A0A0T6AUU9 A0A1B6CFJ0 A0A1B6LGH9 A0A1Q3G2M9 J9JRM5 A0A067QRA5 A0A1B6E0Y9 A0A1B6C195 A0A0V0G891 A0A1B6BZH6 A0A1Q3G2R9 Q17DF2 A0A1Q3FB32 A0A1Q3FAK1 A0A1Q3FA68 A0A0A1XSN1 A0A146LM21 A0A2S2PUU8 E9HBU6 A0A0P5C2Z1 A0A1A9UUE1 A0A1B0ALP4 A0A0P5PTM6 A0A034VNE3 A0A1B0G1D2 A0A1A9Z4P8 A0A0K8V8X4 A0A0P6FUK9 B0X9A4 A0A0P5NAA3 T1JBC4 A0A1A9WRD1 T1PAJ0 A0A0L0CP61 A0A1L8E5J1 A0A0P6EPE1 W8B802 A0A226E0L8 A0A1Q3FB27 A0A0P5MBV0 A0A0P5Q8Y6 A0A1Q3FDH1 A0A1D2MRA9 A0A0A9WBQ3 A0A0P6CZD9 B4L1E4 D6W736 A0A0M4ETI2 A0A069DWN7 A0A2P8XCL1
A0A194PVL5 A0A154P3L6 A0A1Q3G2E6 A0A026WSQ2 A0A336K0Z8 A0A2J7RKI6 A0A1I8P567 A0A0P6GNC7 A0A1B6KR21 A0A1B6LEG6 A0A1B6D8W4 A0A336LQ14 A0A1J1HTC2 A0A1Q3G2H2 A0A158NI67 A0A336K0W4 E0VQJ3 U5ELQ3 E9IB70 A0A1Q3G213 A0A2H8TQM6 A0A1B6DU41 J9JUC2 A0A2S2PLX8 A0A0T6AUU9 A0A1B6CFJ0 A0A1B6LGH9 A0A1Q3G2M9 J9JRM5 A0A067QRA5 A0A1B6E0Y9 A0A1B6C195 A0A0V0G891 A0A1B6BZH6 A0A1Q3G2R9 Q17DF2 A0A1Q3FB32 A0A1Q3FAK1 A0A1Q3FA68 A0A0A1XSN1 A0A146LM21 A0A2S2PUU8 E9HBU6 A0A0P5C2Z1 A0A1A9UUE1 A0A1B0ALP4 A0A0P5PTM6 A0A034VNE3 A0A1B0G1D2 A0A1A9Z4P8 A0A0K8V8X4 A0A0P6FUK9 B0X9A4 A0A0P5NAA3 T1JBC4 A0A1A9WRD1 T1PAJ0 A0A0L0CP61 A0A1L8E5J1 A0A0P6EPE1 W8B802 A0A226E0L8 A0A1Q3FB27 A0A0P5MBV0 A0A0P5Q8Y6 A0A1Q3FDH1 A0A1D2MRA9 A0A0A9WBQ3 A0A0P6CZD9 B4L1E4 D6W736 A0A0M4ETI2 A0A069DWN7 A0A2P8XCL1
Pubmed
EMBL
AB777870
BAM76583.1
AGBW02014532
OWR41503.1
EF092450
ABK92270.2
+ More
BABH01027325 ODYU01000919 SOQ36406.1 GAIX01003658 JAA88902.1 KQ459597 KPI95170.1 KQ434809 KZC06423.1 GFDL01001072 JAV33973.1 KK107111 EZA59013.1 UFQS01000048 UFQT01000048 SSW98596.1 SSX18982.1 NEVH01002717 PNF41358.1 GDIQ01031159 JAN63578.1 GEBQ01026078 JAT13899.1 GEBQ01018073 JAT21904.1 GEDC01015164 JAS22134.1 SSW98595.1 SSX18981.1 CVRI01000020 CRK91245.1 GFDL01001028 JAV34017.1 ADTU01016350 ADTU01016351 SSW98594.1 SSX18980.1 DS235430 EEB15649.1 GANO01001314 JAB58557.1 GL762111 EFZ22144.1 GFDL01001201 JAV33844.1 GFXV01004691 MBW16496.1 GEDC01008102 JAS29196.1 ABLF02028647 ABLF02028649 GGMR01017669 MBY30288.1 LJIG01022763 KRT78849.1 GEDC01025100 GEDC01009938 JAS12198.1 JAS27360.1 GEBQ01017170 JAT22807.1 GFDL01000991 JAV34054.1 ABLF02010982 KK853035 KDR12224.1 GEDC01005710 JAS31588.1 GEDC01030278 JAS07020.1 GECL01001761 JAP04363.1 GEDC01030625 JAS06673.1 GFDL01000927 JAV34118.1 CH477296 EAT44385.1 GFDL01010342 JAV24703.1 GFDL01010465 JAV24580.1 GFDL01010652 JAV24393.1 GBXI01014248 GBXI01000261 JAD00044.1 JAD14031.1 GDHC01010180 JAQ08449.1 GGMS01000104 MBY69307.1 GL732617 EFX70810.1 GDIP01181029 JAJ42373.1 JXJN01000033 JXJN01000034 GDIQ01125921 JAL25805.1 GAKP01014116 GAKP01014114 GAKP01014113 JAC44836.1 CCAG010000051 CCAG010000052 GDHF01029521 GDHF01016998 GDHF01008110 JAI22793.1 JAI35316.1 JAI44204.1 GDIQ01247188 GDIQ01242825 GDIQ01238240 GDIQ01222043 GDIQ01222042 GDIQ01219391 GDIQ01209043 GDIQ01209042 GDIQ01206459 GDIQ01204991 GDIQ01203250 GDIQ01196753 GDIQ01190943 GDIQ01186657 GDIQ01186656 GDIQ01182758 GDIQ01178746 GDIQ01175312 GDIQ01168557 GDIQ01164914 GDIQ01162216 GDIQ01159164 GDIQ01144230 GDIQ01142778 GDIQ01141025 GDIQ01127518 GDIQ01125922 GDIQ01123676 GDIQ01119024 GDIQ01084548 GDIQ01084547 GDIQ01084546 GDIQ01083087 GDIQ01083086 GDIQ01083050 GDIQ01081587 GDIQ01077984 GDIQ01063448 GDIQ01061739 GDIQ01054001 GDIQ01052411 GDIQ01050810 LRGB01002793 JAN40736.1 KZS06312.1 DS232529 EDS43036.1 GDIQ01151957 JAK99768.1 JH432010 KA645714 AFP60343.1 JRES01000127 KNC33967.1 GFDF01000111 JAV13973.1 GDIQ01059749 JAN34988.1 GAMC01011814 GAMC01011813 JAB94742.1 LNIX01000008 OXA50798.1 GFDL01010250 JAV24795.1 GDIQ01162217 JAK89508.1 GDIQ01117532 JAL34194.1 GFDL01009452 JAV25593.1 LJIJ01000641 ODM95639.1 GBHO01038410 GBRD01006961 GDHC01004837 JAG05194.1 JAG58860.1 JAQ13792.1 GDIP01007634 JAM96081.1 CH933810 EDW06665.1 KQ971307 EFA11416.2 CP012528 ALC48876.1 GBGD01000584 JAC88305.1 PYGN01003034 PSN29723.1
BABH01027325 ODYU01000919 SOQ36406.1 GAIX01003658 JAA88902.1 KQ459597 KPI95170.1 KQ434809 KZC06423.1 GFDL01001072 JAV33973.1 KK107111 EZA59013.1 UFQS01000048 UFQT01000048 SSW98596.1 SSX18982.1 NEVH01002717 PNF41358.1 GDIQ01031159 JAN63578.1 GEBQ01026078 JAT13899.1 GEBQ01018073 JAT21904.1 GEDC01015164 JAS22134.1 SSW98595.1 SSX18981.1 CVRI01000020 CRK91245.1 GFDL01001028 JAV34017.1 ADTU01016350 ADTU01016351 SSW98594.1 SSX18980.1 DS235430 EEB15649.1 GANO01001314 JAB58557.1 GL762111 EFZ22144.1 GFDL01001201 JAV33844.1 GFXV01004691 MBW16496.1 GEDC01008102 JAS29196.1 ABLF02028647 ABLF02028649 GGMR01017669 MBY30288.1 LJIG01022763 KRT78849.1 GEDC01025100 GEDC01009938 JAS12198.1 JAS27360.1 GEBQ01017170 JAT22807.1 GFDL01000991 JAV34054.1 ABLF02010982 KK853035 KDR12224.1 GEDC01005710 JAS31588.1 GEDC01030278 JAS07020.1 GECL01001761 JAP04363.1 GEDC01030625 JAS06673.1 GFDL01000927 JAV34118.1 CH477296 EAT44385.1 GFDL01010342 JAV24703.1 GFDL01010465 JAV24580.1 GFDL01010652 JAV24393.1 GBXI01014248 GBXI01000261 JAD00044.1 JAD14031.1 GDHC01010180 JAQ08449.1 GGMS01000104 MBY69307.1 GL732617 EFX70810.1 GDIP01181029 JAJ42373.1 JXJN01000033 JXJN01000034 GDIQ01125921 JAL25805.1 GAKP01014116 GAKP01014114 GAKP01014113 JAC44836.1 CCAG010000051 CCAG010000052 GDHF01029521 GDHF01016998 GDHF01008110 JAI22793.1 JAI35316.1 JAI44204.1 GDIQ01247188 GDIQ01242825 GDIQ01238240 GDIQ01222043 GDIQ01222042 GDIQ01219391 GDIQ01209043 GDIQ01209042 GDIQ01206459 GDIQ01204991 GDIQ01203250 GDIQ01196753 GDIQ01190943 GDIQ01186657 GDIQ01186656 GDIQ01182758 GDIQ01178746 GDIQ01175312 GDIQ01168557 GDIQ01164914 GDIQ01162216 GDIQ01159164 GDIQ01144230 GDIQ01142778 GDIQ01141025 GDIQ01127518 GDIQ01125922 GDIQ01123676 GDIQ01119024 GDIQ01084548 GDIQ01084547 GDIQ01084546 GDIQ01083087 GDIQ01083086 GDIQ01083050 GDIQ01081587 GDIQ01077984 GDIQ01063448 GDIQ01061739 GDIQ01054001 GDIQ01052411 GDIQ01050810 LRGB01002793 JAN40736.1 KZS06312.1 DS232529 EDS43036.1 GDIQ01151957 JAK99768.1 JH432010 KA645714 AFP60343.1 JRES01000127 KNC33967.1 GFDF01000111 JAV13973.1 GDIQ01059749 JAN34988.1 GAMC01011814 GAMC01011813 JAB94742.1 LNIX01000008 OXA50798.1 GFDL01010250 JAV24795.1 GDIQ01162217 JAK89508.1 GDIQ01117532 JAL34194.1 GFDL01009452 JAV25593.1 LJIJ01000641 ODM95639.1 GBHO01038410 GBRD01006961 GDHC01004837 JAG05194.1 JAG58860.1 JAQ13792.1 GDIP01007634 JAM96081.1 CH933810 EDW06665.1 KQ971307 EFA11416.2 CP012528 ALC48876.1 GBGD01000584 JAC88305.1 PYGN01003034 PSN29723.1
Proteomes
UP000007151
UP000005204
UP000053268
UP000076502
UP000053097
UP000235965
+ More
UP000095300 UP000183832 UP000005205 UP000009046 UP000007819 UP000027135 UP000008820 UP000000305 UP000078200 UP000092460 UP000092444 UP000092445 UP000076858 UP000002320 UP000091820 UP000095301 UP000037069 UP000198287 UP000094527 UP000009192 UP000007266 UP000092553 UP000245037
UP000095300 UP000183832 UP000005205 UP000009046 UP000007819 UP000027135 UP000008820 UP000000305 UP000078200 UP000092460 UP000092444 UP000092445 UP000076858 UP000002320 UP000091820 UP000095301 UP000037069 UP000198287 UP000094527 UP000009192 UP000007266 UP000092553 UP000245037
Pfam
PF00225 Kinesin
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
L8B3Q2
A0A212EJ27
A1E131
H9J8D5
A0A2H1V6K0
S4PY99
+ More
A0A194PVL5 A0A154P3L6 A0A1Q3G2E6 A0A026WSQ2 A0A336K0Z8 A0A2J7RKI6 A0A1I8P567 A0A0P6GNC7 A0A1B6KR21 A0A1B6LEG6 A0A1B6D8W4 A0A336LQ14 A0A1J1HTC2 A0A1Q3G2H2 A0A158NI67 A0A336K0W4 E0VQJ3 U5ELQ3 E9IB70 A0A1Q3G213 A0A2H8TQM6 A0A1B6DU41 J9JUC2 A0A2S2PLX8 A0A0T6AUU9 A0A1B6CFJ0 A0A1B6LGH9 A0A1Q3G2M9 J9JRM5 A0A067QRA5 A0A1B6E0Y9 A0A1B6C195 A0A0V0G891 A0A1B6BZH6 A0A1Q3G2R9 Q17DF2 A0A1Q3FB32 A0A1Q3FAK1 A0A1Q3FA68 A0A0A1XSN1 A0A146LM21 A0A2S2PUU8 E9HBU6 A0A0P5C2Z1 A0A1A9UUE1 A0A1B0ALP4 A0A0P5PTM6 A0A034VNE3 A0A1B0G1D2 A0A1A9Z4P8 A0A0K8V8X4 A0A0P6FUK9 B0X9A4 A0A0P5NAA3 T1JBC4 A0A1A9WRD1 T1PAJ0 A0A0L0CP61 A0A1L8E5J1 A0A0P6EPE1 W8B802 A0A226E0L8 A0A1Q3FB27 A0A0P5MBV0 A0A0P5Q8Y6 A0A1Q3FDH1 A0A1D2MRA9 A0A0A9WBQ3 A0A0P6CZD9 B4L1E4 D6W736 A0A0M4ETI2 A0A069DWN7 A0A2P8XCL1
A0A194PVL5 A0A154P3L6 A0A1Q3G2E6 A0A026WSQ2 A0A336K0Z8 A0A2J7RKI6 A0A1I8P567 A0A0P6GNC7 A0A1B6KR21 A0A1B6LEG6 A0A1B6D8W4 A0A336LQ14 A0A1J1HTC2 A0A1Q3G2H2 A0A158NI67 A0A336K0W4 E0VQJ3 U5ELQ3 E9IB70 A0A1Q3G213 A0A2H8TQM6 A0A1B6DU41 J9JUC2 A0A2S2PLX8 A0A0T6AUU9 A0A1B6CFJ0 A0A1B6LGH9 A0A1Q3G2M9 J9JRM5 A0A067QRA5 A0A1B6E0Y9 A0A1B6C195 A0A0V0G891 A0A1B6BZH6 A0A1Q3G2R9 Q17DF2 A0A1Q3FB32 A0A1Q3FAK1 A0A1Q3FA68 A0A0A1XSN1 A0A146LM21 A0A2S2PUU8 E9HBU6 A0A0P5C2Z1 A0A1A9UUE1 A0A1B0ALP4 A0A0P5PTM6 A0A034VNE3 A0A1B0G1D2 A0A1A9Z4P8 A0A0K8V8X4 A0A0P6FUK9 B0X9A4 A0A0P5NAA3 T1JBC4 A0A1A9WRD1 T1PAJ0 A0A0L0CP61 A0A1L8E5J1 A0A0P6EPE1 W8B802 A0A226E0L8 A0A1Q3FB27 A0A0P5MBV0 A0A0P5Q8Y6 A0A1Q3FDH1 A0A1D2MRA9 A0A0A9WBQ3 A0A0P6CZD9 B4L1E4 D6W736 A0A0M4ETI2 A0A069DWN7 A0A2P8XCL1
PDB
6B0L
E-value=0,
Score=1701
Ontologies
GO
GO:0005874
GO:0005524
GO:0008017
GO:0007018
GO:0003777
GO:0016021
GO:0005871
GO:0016887
GO:0005813
GO:0005828
GO:0097431
GO:0090619
GO:0008574
GO:1905515
GO:0007079
GO:0055028
GO:0061673
GO:0051296
GO:0045167
GO:0061867
GO:0070462
GO:0000775
GO:0007057
GO:0098534
GO:0005216
GO:0006811
GO:0045211
GO:0006281
GO:0048384
PANTHER
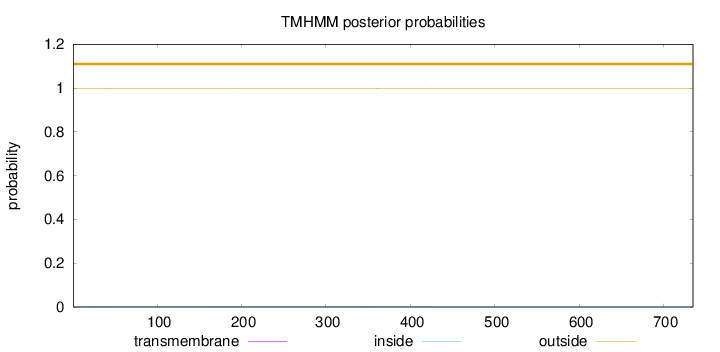
Topology
Length:
735
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05739
Exp number, first 60 AAs:
0.01262
Total prob of N-in:
0.00294
outside
1 - 735
Population Genetic Test Statistics
Pi
31.249121
Theta
26.411803
Tajima's D
0.109124
CLR
1.528302
CSRT
0.406979651017449
Interpretation
Uncertain
