Gene
KWMTBOMO03120 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005793
Annotation
cytochrome_c_oxidase_subunit_Va_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.962
Sequence
CDS
ATGTTGAAATCCGCATCAGGTGCGTTTGCAAATGTCCTTAAAAGATCATTAGTTCCTGCCGTGAAGGTCAGCGCCGTATCTGTACGGAGATCACACGGAGGTCCAGTGGAAAGTGATGAAGAATTCGATAACAGATATGAAGCATATTTCAACAGAAAAGACATTGATGGTTGGGAAATCCGCAAAGGAATGAATGACCTCTGTGGTATGGATCTCGTTCCCGACCCAAAGATTATCAAAGCTGCCCTTCATGCATGCAGACGAGTCAATGATTATGCCCTTGCAGTCAGGTTTATAGAAGCCTGCAAAGACAAATGTGGAAACAAAGTGAATGAAATCTACCCATACATCATTCAAGAGATCAAGCCTACTCTCACCGAACTGGGTATTGACACACCTGAGGAGCTTGGCTACGATAAGCCAGAACTCGCTTTAGAAAGTGTTTACCACATGTAA
Protein
MLKSASGAFANVLKRSLVPAVKVSAVSVRRSHGGPVESDEEFDNRYEAYFNRKDIDGWEIRKGMNDLCGMDLVPDPKIIKAALHACRRVNDYALAVRFIEACKDKCGNKVNEIYPYIIQEIKPTLTELGIDTPEELGYDKPELALESVYHM
Summary
Uniprot
H9J8F1
Q2F5S6
A0A2A4JDL8
E7EC46
A0A2W1BJ66
A0A2H1VMS2
+ More
A0A3G1T1P2 A0A0K8SHA2 A0A0N1INM6 I4DJ18 A0A0L7LKX5 I4DMK7 A0A1E1W6Z6 S4PE47 A0A1Q3FL80 T1D575 Q1HRJ0 A0A023EGZ5 B0WL66 A0A1Y1MUG9 D7EJQ8 A0A182T5X2 A0A182RTD2 V5IAE1 A0A1Y9IWA2 Q95V82 A0A1W4WZR1 A0A182MTZ5 A0A240PP78 A0A182NZM2 A0A2P8YFR3 A0A1Y9J0C1 A0A1Y9IRS7 A0A1C7CZB9 A0A1I8JUX6 A0A1Y9GK70 Q7PSR9 A0A2J7R3H8 A0A1Y9H9I7 A0A182NW86 M1FZ15 D1FQE8 A0A084VHT6 A0A2M3ZFU9 R4UK26 A0A1Y1NJE5 A0A240PP05 W5J7I0 A0A182FKK7 A0A2M4A5U6 A0A0K8TTZ3 A0A0P4WGT0 A0A0A9XB66 A0A067RQ95 A0A1B0GMS7 A0A146LDL7 T1DER5 A0A348G665 A0A2R7VQF9 A0A0L8FZR5 W8BY83 E2CAG7 A0A293M7U3 U5EVZ7 A0A1L8E0L7 A0A293MKF2 A0A195F1K2 A0A0N8CEL7 A0A2R5LFV7 A0A0P5Z3D8 A0A151XGD2 A0A0P5HNK7 T1II41 A0A151JRH0 A0A023FCS1 A6YPL2 A0A0P5ALC9 A0A034WA24 A0A0C9RH34 A0A0P5Z8T7 A0A034W8F5 A0A131Y626 Q09JF2 A0A0V0G3K0 A0A1J1IFV2 A0A0P4Z405 A0A0N7Z9M0 R4G8K2 A0A171AJR0 A0A131Y3G2 A0A0K8RJ43 B7Q1Q0 F4X159 Q6B867 B2D2D5 A0A023FHX2 Q16Q43 K7IUM0 A0A0C9R012 A0A224Y0F8
A0A3G1T1P2 A0A0K8SHA2 A0A0N1INM6 I4DJ18 A0A0L7LKX5 I4DMK7 A0A1E1W6Z6 S4PE47 A0A1Q3FL80 T1D575 Q1HRJ0 A0A023EGZ5 B0WL66 A0A1Y1MUG9 D7EJQ8 A0A182T5X2 A0A182RTD2 V5IAE1 A0A1Y9IWA2 Q95V82 A0A1W4WZR1 A0A182MTZ5 A0A240PP78 A0A182NZM2 A0A2P8YFR3 A0A1Y9J0C1 A0A1Y9IRS7 A0A1C7CZB9 A0A1I8JUX6 A0A1Y9GK70 Q7PSR9 A0A2J7R3H8 A0A1Y9H9I7 A0A182NW86 M1FZ15 D1FQE8 A0A084VHT6 A0A2M3ZFU9 R4UK26 A0A1Y1NJE5 A0A240PP05 W5J7I0 A0A182FKK7 A0A2M4A5U6 A0A0K8TTZ3 A0A0P4WGT0 A0A0A9XB66 A0A067RQ95 A0A1B0GMS7 A0A146LDL7 T1DER5 A0A348G665 A0A2R7VQF9 A0A0L8FZR5 W8BY83 E2CAG7 A0A293M7U3 U5EVZ7 A0A1L8E0L7 A0A293MKF2 A0A195F1K2 A0A0N8CEL7 A0A2R5LFV7 A0A0P5Z3D8 A0A151XGD2 A0A0P5HNK7 T1II41 A0A151JRH0 A0A023FCS1 A6YPL2 A0A0P5ALC9 A0A034WA24 A0A0C9RH34 A0A0P5Z8T7 A0A034W8F5 A0A131Y626 Q09JF2 A0A0V0G3K0 A0A1J1IFV2 A0A0P4Z405 A0A0N7Z9M0 R4G8K2 A0A171AJR0 A0A131Y3G2 A0A0K8RJ43 B7Q1Q0 F4X159 Q6B867 B2D2D5 A0A023FHX2 Q16Q43 K7IUM0 A0A0C9R012 A0A224Y0F8
Pubmed
19121390
28756777
26354079
22651552
26227816
23622113
+ More
24330624 17204158 17510324 24945155 26483478 28004739 18362917 19820115 14518003 29403074 20966253 12364791 14747013 17210077 23831752 24438588 20920257 23761445 26369729 25401762 24845553 26823975 24495485 20798317 25474469 18207082 25348373 18070664 27129103 21719571 16102420 18725333 20075255 26131772
24330624 17204158 17510324 24945155 26483478 28004739 18362917 19820115 14518003 29403074 20966253 12364791 14747013 17210077 23831752 24438588 20920257 23761445 26369729 25401762 24845553 26823975 24495485 20798317 25474469 18207082 25348373 18070664 27129103 21719571 16102420 18725333 20075255 26131772
EMBL
BABH01027330
DQ311347
ABD36291.1
NWSH01001805
PCG70071.1
HQ649756
+ More
JN082712 ADV36662.1 AER27810.1 KZ150156 PZC72770.1 ODYU01003405 SOQ42129.1 MG992457 AXY94895.1 GBRD01013666 JAG52160.1 KQ460970 KPJ10189.1 AK401286 KQ459597 BAM17908.1 KPI95167.1 JTDY01000791 KOB75861.1 AK402525 BAM19147.1 GDQN01008330 JAT82724.1 GAIX01003326 JAA89234.1 GFDL01006781 JAV28264.1 GALA01000658 JAA94194.1 DQ017468 DQ440104 CH478532 AAY41836.1 ABF18137.1 EAT32822.1 JXUM01146375 GAPW01005473 KQ570161 JAC08125.1 KXJ68438.1 DS231980 EDS30239.1 GEZM01026377 JAV87027.1 DS497711 EFA12825.1 GALX01001345 JAB67121.1 AF420467 AAL17607.1 AXCM01001359 PYGN01000632 PSN43071.1 APCN01001484 AAAB01008816 EAA05128.4 NEVH01007819 PNF35376.1 AXCN02001729 JQ839284 AFW19996.1 EZ420048 ACZ28403.1 ATLV01013235 KE524847 KFB37530.1 GGFM01006628 MBW27379.1 KC571935 AGM32434.1 GEZM01003500 JAV96765.1 ADMH02002090 ETN59358.1 GGFK01002842 MBW36163.1 GDAI01000173 JAI17430.1 GDRN01054129 JAI66118.1 GBHO01026385 GBRD01007899 JAG17219.1 JAG57922.1 KK852486 KDR22790.1 AJVK01012257 GDHC01014019 JAQ04610.1 GAMD01003426 JAA98164.1 FX985607 BBF97938.1 KK854027 PTY09754.1 KQ425021 KOF70079.1 GAMC01002243 GAMC01002241 JAC04313.1 GL453976 EFN75078.1 GFWV01010296 MAA35025.1 GANO01000775 JAB59096.1 GFDF01001972 JAV12112.1 GFWV01015679 MAA40408.1 KQ981880 KYN33974.1 GDIP01139311 JAL64403.1 GGLE01004233 MBY08359.1 GDIP01050524 JAM53191.1 KQ982174 KYQ59375.1 GDIQ01225519 JAK26206.1 JH430102 KQ978589 KYN29969.1 GBBI01000079 JAC18633.1 EF639023 ABR27908.1 GDIP01200604 JAJ22798.1 GAKP01007962 JAC50990.1 GBYB01007625 JAG77392.1 GDIP01048427 LRGB01002451 JAM55288.1 KZS07460.1 GAKP01007963 JAC50989.1 GEFM01001861 JAP73935.1 DQ886894 ABI52811.1 GECL01003468 JAP02656.1 CVRI01000049 CRK99143.1 GDIP01230405 JAI92996.1 GDKW01000171 JAI56424.1 ACPB03009273 GAHY01000827 JAA76683.1 GEMB01000984 JAS02160.1 GEFM01001763 JAP74033.1 GADI01003244 JAA70564.1 ABJB010083726 ABJB010874727 DS838808 EEC12772.1 GL888526 EGI59826.1 AY674281 AAT92214.1 EU574869 ACB70376.1 GBBK01004119 JAC20363.1 CH477762 EAT36510.1 AAZX01002642 GBZX01002400 JAG90340.1 GFTR01002545 JAW13881.1
JN082712 ADV36662.1 AER27810.1 KZ150156 PZC72770.1 ODYU01003405 SOQ42129.1 MG992457 AXY94895.1 GBRD01013666 JAG52160.1 KQ460970 KPJ10189.1 AK401286 KQ459597 BAM17908.1 KPI95167.1 JTDY01000791 KOB75861.1 AK402525 BAM19147.1 GDQN01008330 JAT82724.1 GAIX01003326 JAA89234.1 GFDL01006781 JAV28264.1 GALA01000658 JAA94194.1 DQ017468 DQ440104 CH478532 AAY41836.1 ABF18137.1 EAT32822.1 JXUM01146375 GAPW01005473 KQ570161 JAC08125.1 KXJ68438.1 DS231980 EDS30239.1 GEZM01026377 JAV87027.1 DS497711 EFA12825.1 GALX01001345 JAB67121.1 AF420467 AAL17607.1 AXCM01001359 PYGN01000632 PSN43071.1 APCN01001484 AAAB01008816 EAA05128.4 NEVH01007819 PNF35376.1 AXCN02001729 JQ839284 AFW19996.1 EZ420048 ACZ28403.1 ATLV01013235 KE524847 KFB37530.1 GGFM01006628 MBW27379.1 KC571935 AGM32434.1 GEZM01003500 JAV96765.1 ADMH02002090 ETN59358.1 GGFK01002842 MBW36163.1 GDAI01000173 JAI17430.1 GDRN01054129 JAI66118.1 GBHO01026385 GBRD01007899 JAG17219.1 JAG57922.1 KK852486 KDR22790.1 AJVK01012257 GDHC01014019 JAQ04610.1 GAMD01003426 JAA98164.1 FX985607 BBF97938.1 KK854027 PTY09754.1 KQ425021 KOF70079.1 GAMC01002243 GAMC01002241 JAC04313.1 GL453976 EFN75078.1 GFWV01010296 MAA35025.1 GANO01000775 JAB59096.1 GFDF01001972 JAV12112.1 GFWV01015679 MAA40408.1 KQ981880 KYN33974.1 GDIP01139311 JAL64403.1 GGLE01004233 MBY08359.1 GDIP01050524 JAM53191.1 KQ982174 KYQ59375.1 GDIQ01225519 JAK26206.1 JH430102 KQ978589 KYN29969.1 GBBI01000079 JAC18633.1 EF639023 ABR27908.1 GDIP01200604 JAJ22798.1 GAKP01007962 JAC50990.1 GBYB01007625 JAG77392.1 GDIP01048427 LRGB01002451 JAM55288.1 KZS07460.1 GAKP01007963 JAC50989.1 GEFM01001861 JAP73935.1 DQ886894 ABI52811.1 GECL01003468 JAP02656.1 CVRI01000049 CRK99143.1 GDIP01230405 JAI92996.1 GDKW01000171 JAI56424.1 ACPB03009273 GAHY01000827 JAA76683.1 GEMB01000984 JAS02160.1 GEFM01001763 JAP74033.1 GADI01003244 JAA70564.1 ABJB010083726 ABJB010874727 DS838808 EEC12772.1 GL888526 EGI59826.1 AY674281 AAT92214.1 EU574869 ACB70376.1 GBBK01004119 JAC20363.1 CH477762 EAT36510.1 AAZX01002642 GBZX01002400 JAG90340.1 GFTR01002545 JAW13881.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000008820
+ More
UP000069940 UP000249989 UP000002320 UP000007266 UP000075901 UP000075900 UP000075920 UP000192223 UP000075883 UP000075881 UP000075885 UP000245037 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000235965 UP000075886 UP000075884 UP000030765 UP000075880 UP000000673 UP000069272 UP000027135 UP000092462 UP000053454 UP000008237 UP000078541 UP000075809 UP000078492 UP000076858 UP000183832 UP000015103 UP000001555 UP000007755 UP000002358
UP000069940 UP000249989 UP000002320 UP000007266 UP000075901 UP000075900 UP000075920 UP000192223 UP000075883 UP000075881 UP000075885 UP000245037 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000235965 UP000075886 UP000075884 UP000030765 UP000075880 UP000000673 UP000069272 UP000027135 UP000092462 UP000053454 UP000008237 UP000078541 UP000075809 UP000078492 UP000076858 UP000183832 UP000015103 UP000001555 UP000007755 UP000002358
Pfam
PF02284 COX5A
SUPFAM
SSF48479
SSF48479
Gene 3D
ProteinModelPortal
H9J8F1
Q2F5S6
A0A2A4JDL8
E7EC46
A0A2W1BJ66
A0A2H1VMS2
+ More
A0A3G1T1P2 A0A0K8SHA2 A0A0N1INM6 I4DJ18 A0A0L7LKX5 I4DMK7 A0A1E1W6Z6 S4PE47 A0A1Q3FL80 T1D575 Q1HRJ0 A0A023EGZ5 B0WL66 A0A1Y1MUG9 D7EJQ8 A0A182T5X2 A0A182RTD2 V5IAE1 A0A1Y9IWA2 Q95V82 A0A1W4WZR1 A0A182MTZ5 A0A240PP78 A0A182NZM2 A0A2P8YFR3 A0A1Y9J0C1 A0A1Y9IRS7 A0A1C7CZB9 A0A1I8JUX6 A0A1Y9GK70 Q7PSR9 A0A2J7R3H8 A0A1Y9H9I7 A0A182NW86 M1FZ15 D1FQE8 A0A084VHT6 A0A2M3ZFU9 R4UK26 A0A1Y1NJE5 A0A240PP05 W5J7I0 A0A182FKK7 A0A2M4A5U6 A0A0K8TTZ3 A0A0P4WGT0 A0A0A9XB66 A0A067RQ95 A0A1B0GMS7 A0A146LDL7 T1DER5 A0A348G665 A0A2R7VQF9 A0A0L8FZR5 W8BY83 E2CAG7 A0A293M7U3 U5EVZ7 A0A1L8E0L7 A0A293MKF2 A0A195F1K2 A0A0N8CEL7 A0A2R5LFV7 A0A0P5Z3D8 A0A151XGD2 A0A0P5HNK7 T1II41 A0A151JRH0 A0A023FCS1 A6YPL2 A0A0P5ALC9 A0A034WA24 A0A0C9RH34 A0A0P5Z8T7 A0A034W8F5 A0A131Y626 Q09JF2 A0A0V0G3K0 A0A1J1IFV2 A0A0P4Z405 A0A0N7Z9M0 R4G8K2 A0A171AJR0 A0A131Y3G2 A0A0K8RJ43 B7Q1Q0 F4X159 Q6B867 B2D2D5 A0A023FHX2 Q16Q43 K7IUM0 A0A0C9R012 A0A224Y0F8
A0A3G1T1P2 A0A0K8SHA2 A0A0N1INM6 I4DJ18 A0A0L7LKX5 I4DMK7 A0A1E1W6Z6 S4PE47 A0A1Q3FL80 T1D575 Q1HRJ0 A0A023EGZ5 B0WL66 A0A1Y1MUG9 D7EJQ8 A0A182T5X2 A0A182RTD2 V5IAE1 A0A1Y9IWA2 Q95V82 A0A1W4WZR1 A0A182MTZ5 A0A240PP78 A0A182NZM2 A0A2P8YFR3 A0A1Y9J0C1 A0A1Y9IRS7 A0A1C7CZB9 A0A1I8JUX6 A0A1Y9GK70 Q7PSR9 A0A2J7R3H8 A0A1Y9H9I7 A0A182NW86 M1FZ15 D1FQE8 A0A084VHT6 A0A2M3ZFU9 R4UK26 A0A1Y1NJE5 A0A240PP05 W5J7I0 A0A182FKK7 A0A2M4A5U6 A0A0K8TTZ3 A0A0P4WGT0 A0A0A9XB66 A0A067RQ95 A0A1B0GMS7 A0A146LDL7 T1DER5 A0A348G665 A0A2R7VQF9 A0A0L8FZR5 W8BY83 E2CAG7 A0A293M7U3 U5EVZ7 A0A1L8E0L7 A0A293MKF2 A0A195F1K2 A0A0N8CEL7 A0A2R5LFV7 A0A0P5Z3D8 A0A151XGD2 A0A0P5HNK7 T1II41 A0A151JRH0 A0A023FCS1 A6YPL2 A0A0P5ALC9 A0A034WA24 A0A0C9RH34 A0A0P5Z8T7 A0A034W8F5 A0A131Y626 Q09JF2 A0A0V0G3K0 A0A1J1IFV2 A0A0P4Z405 A0A0N7Z9M0 R4G8K2 A0A171AJR0 A0A131Y3G2 A0A0K8RJ43 B7Q1Q0 F4X159 Q6B867 B2D2D5 A0A023FHX2 Q16Q43 K7IUM0 A0A0C9R012 A0A224Y0F8
PDB
2Y69
E-value=2.67718e-37,
Score=383
Ontologies
PATHWAY
GO
PANTHER
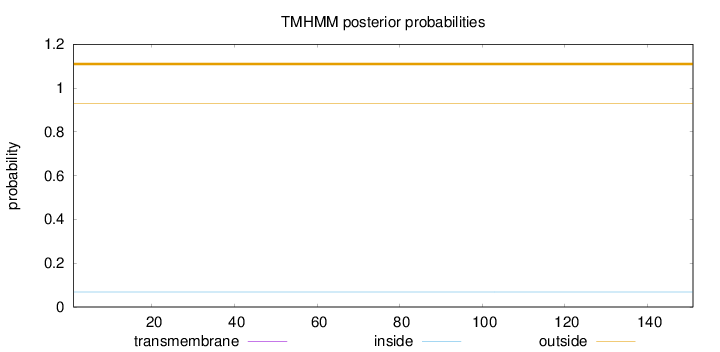
Topology
Length:
151
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00078
Exp number, first 60 AAs:
0.00078
Total prob of N-in:
0.06898
outside
1 - 151
Population Genetic Test Statistics
Pi
14.37324
Theta
17.910088
Tajima's D
0.614415
CLR
4.511592
CSRT
0.561821908904555
Interpretation
Uncertain
