Gene
KWMTBOMO03116 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005772
Annotation
neuroglian_[Mythimna_separata]
Full name
Neuroglian
Location in the cell
Cytoplasmic Reliability : 1.05 Extracellular Reliability : 1.055
Sequence
CDS
ATGGATACGAGCAGAATTCTGTGTTTCATAGCGTTAGTCTCGCAGGCCGCTGCTTTATTAACATCTCCACCAAAAATCGTGAAGCAACCGACTCAGGAAGAAATCCTGTTCCAAGTGGCGCAACAGGGAGAAGTGGATAAACCTTTTATTATAGAGTGCGAGGCTGAAGGAGAACCAGCACCTAAGTACAAATGGATCAAAAACGGCAAATCATTCGAATACTCGAGCTACGACAACCGTATCTCGCAACAAACCGGTCGAGGTACTCTCGTCATCAGCAAGCCCAAGGAGGAAGATCTGGGACAGTACCAGTGCTTCGCCTACAACGAGTGGGGAACCGCTGCATCCAACTCGGTCTTTGTAAGGAGAGCCGAGCTCAATTCCTTCAAGGAAACCGATGGTCAGCAAGTTGTGAGAGCTGAAGAAGGTAAACCTTATAAGCTGACGTGCGAGCCCCCGGACGGTCATCCGAAGCCCAAGGTCTACTGGATGTTGCAAGGGGACCAGGGTCAGCTGAAGACGATCAACAACTCTCGTATGACACTCGACCCGGAAGGAAACCTCTGGTTCTCCAACGTGACCCAAAACGACGCCAGCGAAGATTACGCGTACATCTGCACGGCCAACTCCGGCTTCAGAAATGAATACAAAGTTGGGAACAAGATCTACCTTCAGGTCATCCCGACCGGCATCTCGCCGACACTCAACCGACACGAGCCTGTGCGCCAGTACACCACCAGGAAGATCGAGAGAGGCCTCAAGGGTCAGCGCGTGGAGCTCTACTGCATCTACGGCGGAACGCCGCTCCCTCAGATCGTATGGAAGAAGAACGGGCGCAGCATCTTGTGGTCTCAGAGCATCACCCAGGACAACTACGGGAAGACTTTAGTGATCAAGTATCCGACGTACGACGACCAGGGCACGTACACGTGCGAGGTCAGCAACGGTGTGGGCTCGGCGCAGTCCAACTCCATCCAACTCAACATCGAAGCTGCGCCATTCTTCACGGTGGAACCAGAGATCCAGAACCTAGCCGAGGGCGAGACCGCGGTCATCCGCTGCGAGGCGGACGGCACCCCGGCGCCGAAGATATCCTGGATCCACAACGGGAAGCCGATCGAGCAGGCCGAGCTGAACCCGCGCCGCCAAGTCAGCGCCAACTCCATCGTCATCACCGACCTCGTGAAGAAGGACACGGGCAACTACGGCTGCAACGCCACCAGCGCCATCGGCTACGTCTACAAGGACGTCTACATCAACGTGCAGTCCATTCCCCCTGAAATCAAGGAGGGTCCCATGAACCTGACCAACGTGGACGGCTCCGAGGCTGTGCTCAAGTGCAGAGTCTTTGGAGCACCGAAACCTATCGTTAAATGGATGAAGGACAACGTCGACGTCACAGGAGGGAAATACAACATAACGGCGGAGGGCGACCTCGTGATCCGCGACGTGTCGTTCACTGACGTGGGCACGTACCAGTGCTACGCCAAGAACAAGTTCGGCGAGACCTCCGCCTACGGATCGCTGGTGGTCAAGAAGCACACGGTGATCACGGACAAGCCGGAGCACTACGAGGTGGCGGCGGGCTCGTCGGCCACGTTCCGCTGCAACGCCAACGCGGACGACTCGCTGCCGCTGCAAATCATCTGGCTCAACGACGGCCAGCAGATCGACTTCGAGAGCCAGCCGCGCTTCCGCATGACCAACGACTACTCGCTGCTCATCTCCGACACCAGCGAGCTCGACTCCGGCCAGTACACCTGCATCGCCAGGACGCCCGTCGACGAGGCGCGCGCCCAGGCCACGCTCACCGTCCAAGGGTGTGGAGCTTGGTATTTGAGTGTGAGTGTGTGTGCAGACAAGCCGAACCCGCCGGTGCTGACGGGCGCGGAGTGCGGCGCGCGCACGGCCACGGTGCGGTGGCGGGCGGCGGGCGACAACCGCGCGCCCATCCTGCGCTACTCGCTGCACTACAACACCAGCTTCACGCCGCACGTGTGGGCCGTCGCCGCCGCCGCCGTGCCGCCAGTAGACTCCGCCTGGACCGCCGCGCTCAGCCCCTGGGCCAACTACACCTTCCGCGTGGTGGCGCACAACAAGATCGGCCCGTCAGCGCCCTCGGGCCACTCCGACGTGTGCACCACGCAGCCCGACGTGCCCTACAAGAACCCCGACAACGTGAAGGGCGAGGGCACCGACCCCACCAACATGCTCATATCGTGGACGAAAATGCCTCAAATCGAACACAACGGCCCCGGGTTCTACTATCTGGTGTCGTGGCGCCGGAACATCACGGGACAACCCTGGGAAGAGCACCAGGTCCGGGACTGGCAGCAAACCGAACACCTGGTGACCAACACCCCCACCTTCCAGCCCTACAAGATCAAGGTGACGGCGGTGAACTTCAAAGGAACATCGAACGTGACTCCCATTGAAGTGATCGGCTGGTCCGGAGAGGACCGACCCACTCAGGCTCCGGGCAACCTCAGCCTGTCCCAGGTGACGTCCGGCACCAGCGCCGTGCTCAGCTGGACTCCCGTGGCGCCGGAGTCACTGCGGGGGCACTTCAAAGGATACAAAATCCAGACCTGGACCGACGCCGAGGAGGACCGGCTCAAGGAGATCTTTGTGGAGTCCGACGCGACCAGCGCCCTCGTCAACAAGTTCAAGCCCTTCAAGAAGAACAACGTGCGCATCCTCGCCTACAACGGCCGCTTCAACGGGCCCGCCAGCGACACCCTCAGCTTCGTCACCCCCGAGGGACAGCCCTCCACCGTTCGCTCCTTCGAGGCCTACCCTATCGGCTCCTCCGCCATGCTGCTCAAGTGGGAGAAGCCCATGGAGGAGAACGGCGTCCTCACCGGCTACAAGATCTACTACAGCAAGGTGATCGCCACGGCCGTGGAGACTCCGAAGGAGCGCAAGAAGGAGATCGACCCCAAGTTCGACCGCGCCAAGCTCGCCGGCCTCGAGCCCAACACCAAGTACAGGATCGAGATCCGCGCCAAGACCAAGGCCGGCGAGGGCAAGGGCTACTACGTGGAGCAGTCCACCAACAACGTGCTCACCGCGCAGCCCGACGTGCCCGCCTTCGAGACCGAGACGCTGCCCGGGAAGGAGGGCACGGCCCACGTCATAGTCCGCTGGATCCCCTCGCTCGACGGACACGCGGGGACGCACTTCGTGGCCTGGTACCGGCTCAAGGGGCACCCCGACTGGCAGCGGACCAACGACATCACCGAGGACGACTTCGTGATCCTGACGGGGCTGGAGCCCGGCCAGCTGTACGAGGTCAAGGTCACGGTGCACGACGGCCACTACTTCAGCACCAGCGAGCTGCGGACGGTCGACACCTCCATAGTAGACGGACCGATAGTGAAGCCGGACGAGAAGATCGCGACCGCGGGCTGGTTCATCGGCGTGATGCTGGCGCTGGCCTTCCTGCTGCTGCTGCTGGTGCTGGTGTGCGTGCTGCGCAGGAACCGCGGCGGCAAGTACGACGTGCACGACCGCGAGCTGGCGCACGGCCGCAGGGACTACCCCGACGCCGGCTTCCACGAGTACACGCACCCATTGGACAACAAGTCTCGCCATTCGATGAGCAGCGGCACCAAGCCGGGGCCCGAGAGCGACACGGACTCGATGGCGGAGTACGGCGACGGAGAGACGGCCGGCATGAACGAGGACGGCTCCTTCATCGGACAGTACGGCCGGAAACGACGTCCGCCAACATCCGGCCAGCCGGACTCAGCCTCGCAAGCATTTGCAACGCTCGTTTAA
Protein
MDTSRILCFIALVSQAAALLTSPPKIVKQPTQEEILFQVAQQGEVDKPFIIECEAEGEPAPKYKWIKNGKSFEYSSYDNRISQQTGRGTLVISKPKEEDLGQYQCFAYNEWGTAASNSVFVRRAELNSFKETDGQQVVRAEEGKPYKLTCEPPDGHPKPKVYWMLQGDQGQLKTINNSRMTLDPEGNLWFSNVTQNDASEDYAYICTANSGFRNEYKVGNKIYLQVIPTGISPTLNRHEPVRQYTTRKIERGLKGQRVELYCIYGGTPLPQIVWKKNGRSILWSQSITQDNYGKTLVIKYPTYDDQGTYTCEVSNGVGSAQSNSIQLNIEAAPFFTVEPEIQNLAEGETAVIRCEADGTPAPKISWIHNGKPIEQAELNPRRQVSANSIVITDLVKKDTGNYGCNATSAIGYVYKDVYINVQSIPPEIKEGPMNLTNVDGSEAVLKCRVFGAPKPIVKWMKDNVDVTGGKYNITAEGDLVIRDVSFTDVGTYQCYAKNKFGETSAYGSLVVKKHTVITDKPEHYEVAAGSSATFRCNANADDSLPLQIIWLNDGQQIDFESQPRFRMTNDYSLLISDTSELDSGQYTCIARTPVDEARAQATLTVQGCGAWYLSVSVCADKPNPPVLTGAECGARTATVRWRAAGDNRAPILRYSLHYNTSFTPHVWAVAAAAVPPVDSAWTAALSPWANYTFRVVAHNKIGPSAPSGHSDVCTTQPDVPYKNPDNVKGEGTDPTNMLISWTKMPQIEHNGPGFYYLVSWRRNITGQPWEEHQVRDWQQTEHLVTNTPTFQPYKIKVTAVNFKGTSNVTPIEVIGWSGEDRPTQAPGNLSLSQVTSGTSAVLSWTPVAPESLRGHFKGYKIQTWTDAEEDRLKEIFVESDATSALVNKFKPFKKNNVRILAYNGRFNGPASDTLSFVTPEGQPSTVRSFEAYPIGSSAMLLKWEKPMEENGVLTGYKIYYSKVIATAVETPKERKKEIDPKFDRAKLAGLEPNTKYRIEIRAKTKAGEGKGYYVEQSTNNVLTAQPDVPAFETETLPGKEGTAHVIVRWIPSLDGHAGTHFVAWYRLKGHPDWQRTNDITEDDFVILTGLEPGQLYEVKVTVHDGHYFSTSELRTVDTSIVDGPIVKPDEKIATAGWFIGVMLALAFLLLLLVLVCVLRRNRGGKYDVHDRELAHGRRDYPDAGFHEYTHPLDNKSRHSMSSGTKPGPESDTDSMAEYGDGETAGMNEDGSFIGQYGRKRRPPTSGQPDSASQAFATLV
Summary
Description
The long isoform may play a role in neural and glial cell adhesion in the developing embryo. The short isoform may be a more general cell adhesion molecule involved in other tissues and imaginal disk morphogenesis. Vital for embryonic development. Essential for septate junctions. Septate junctions, which are the equivalent of vertebrates tight junctions, are characterized by regular arrays of transverse structures that span the intermembrane space and form a physical barrier to diffusion. Required for the blood-brain barrier formation.
Subunit
Forms a complex with Nrx and Cont.
Keywords
3D-structure
Alternative splicing
Cell adhesion
Cell junction
Cell membrane
Complete proteome
Developmental protein
Direct protein sequencing
Disulfide bond
Glycoprotein
Immunoglobulin domain
Membrane
Reference proteome
Repeat
Signal
Tight junction
Transmembrane
Transmembrane helix
Feature
chain Neuroglian
splice variant In isoform A.
splice variant In isoform A.
Uniprot
D0VY44
A0A2W1BGE0
A0A2H1VMQ7
P91767
A0A212EQR2
A0A194PPC6
+ More
A0A0N1IE92 A0A1B6K0I8 A0A1L8DM81 A0A2J7PJ24 U5EU82 A0A2J7PJ23 A0A2J7PJ28 E0VWB3 A0A0A9Z9I6 A0A0A9Y9L8 A0A0L0C8N9 A0A1Q3FJR5 A0A1Q3FJL7 A0A1Q3FJH2 A0A1Q3FJF3 A0A1Q3FJP3 A0A0A9YFP0 A0A0V0G851 A0A0A9ZCK9 A0A1W4VUK4 A0A182V2N1 A0A182X573 A0A0Q9WUV5 B3NVG5 Q7PG72 P20241-2 A0A1W4VGI6 A0A2A3EQC2 B4NCI8 E1JJF9 A0A0Q5T3I4 A0A1I8M4Q6 B4Q0A3 A0A0R3P7J0 E1JJF8 P20241 Q29FM1 A0A0P8ZS96 A0A0R1EHH6 A0A3B0JXM4 A0A1I8PSB9 A0A3B0J8C5 B3N0B0 A0A1I8PS80 A0A182MZQ2 A0A224XGR9 A0A1Q3FJG2 A0A084VVR8 A0A1Q3FJI5 B4HA25 A0A088A0C7 A0A336L9G0 A0A0Q9X0E4 A0A2M3YYL2 F5HJW6 A0A0L7R1U9 A0A0A1XPS4 A0A2M4BA85 B4L315 W8ARL3 W5J7Y4 A0A154P2X8 B4JP14 A0A0Q5THL2 A0A0N1IT65 M9NEZ5 A0A2M3YYL5 A0A2M3YYM0 A0A2M4A1U8 A0A1W4X2R0 B4M1M9 A0A0R1EHL1 A0A2M4BA33 A0A158P3P7 A0A0K8WB81 A0A0K8W814 A0A2M4BA73 A0A2M4A1U2 A0A034V7N8 A0A3B0J7M4 A0A2M3YYJ9 A0A1A9V6L8 B4IJX8 A0A1A9WVD0 A0A0M5J3D9 A0A1B0G439 A0A1B0AXR8 A0A1B0GMP2 A0A151XGE0 A0A182FML2 Q16Z16 K7IUL4 A0A0Q9WVH7 A0A336MRR8
A0A0N1IE92 A0A1B6K0I8 A0A1L8DM81 A0A2J7PJ24 U5EU82 A0A2J7PJ23 A0A2J7PJ28 E0VWB3 A0A0A9Z9I6 A0A0A9Y9L8 A0A0L0C8N9 A0A1Q3FJR5 A0A1Q3FJL7 A0A1Q3FJH2 A0A1Q3FJF3 A0A1Q3FJP3 A0A0A9YFP0 A0A0V0G851 A0A0A9ZCK9 A0A1W4VUK4 A0A182V2N1 A0A182X573 A0A0Q9WUV5 B3NVG5 Q7PG72 P20241-2 A0A1W4VGI6 A0A2A3EQC2 B4NCI8 E1JJF9 A0A0Q5T3I4 A0A1I8M4Q6 B4Q0A3 A0A0R3P7J0 E1JJF8 P20241 Q29FM1 A0A0P8ZS96 A0A0R1EHH6 A0A3B0JXM4 A0A1I8PSB9 A0A3B0J8C5 B3N0B0 A0A1I8PS80 A0A182MZQ2 A0A224XGR9 A0A1Q3FJG2 A0A084VVR8 A0A1Q3FJI5 B4HA25 A0A088A0C7 A0A336L9G0 A0A0Q9X0E4 A0A2M3YYL2 F5HJW6 A0A0L7R1U9 A0A0A1XPS4 A0A2M4BA85 B4L315 W8ARL3 W5J7Y4 A0A154P2X8 B4JP14 A0A0Q5THL2 A0A0N1IT65 M9NEZ5 A0A2M3YYL5 A0A2M3YYM0 A0A2M4A1U8 A0A1W4X2R0 B4M1M9 A0A0R1EHL1 A0A2M4BA33 A0A158P3P7 A0A0K8WB81 A0A0K8W814 A0A2M4BA73 A0A2M4A1U2 A0A034V7N8 A0A3B0J7M4 A0A2M3YYJ9 A0A1A9V6L8 B4IJX8 A0A1A9WVD0 A0A0M5J3D9 A0A1B0G439 A0A1B0AXR8 A0A1B0GMP2 A0A151XGE0 A0A182FML2 Q16Z16 K7IUL4 A0A0Q9WVH7 A0A336MRR8
Pubmed
28756777
9015260
22118469
26354079
20566863
25401762
+ More
26823975 26108605 17994087 12364791 14747013 17210077 2805067 9666073 10731132 12537572 12537569 1693086 15459097 17893096 19349973 7512815 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 15632085 23185243 18057021 24438588 25830018 24495485 20920257 23761445 21347285 25348373 17510324 20075255
26823975 26108605 17994087 12364791 14747013 17210077 2805067 9666073 10731132 12537572 12537569 1693086 15459097 17893096 19349973 7512815 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 15632085 23185243 18057021 24438588 25830018 24495485 20920257 23761445 21347285 25348373 17510324 20075255
EMBL
AB490501
BAI49425.1
KZ150156
PZC72775.1
ODYU01003405
SOQ42125.1
+ More
U50719 AAC47451.1 AGBW02013246 OWR43836.1 KQ459597 KPI95162.1 KQ460970 KPJ10193.1 GECU01002740 JAT04967.1 GFDF01006522 JAV07562.1 NEVH01024950 PNF16340.1 GANO01002435 JAB57436.1 PNF16341.1 PNF16342.1 DS235818 EEB17669.1 GBHO01002530 GBRD01018117 GDHC01017353 GDHC01003765 JAG41074.1 JAG47710.1 JAQ01276.1 JAQ14864.1 GBHO01015288 GBRD01017682 GDHC01016665 JAG28316.1 JAG48145.1 JAQ01964.1 JRES01000755 KNC28615.1 GFDL01007303 JAV27742.1 GFDL01007330 JAV27715.1 GFDL01007275 JAV27770.1 GFDL01007295 JAV27750.1 GFDL01007323 JAV27722.1 GBHO01015279 JAG28325.1 GECL01001809 JAP04315.1 GBHO01002526 JAG41078.1 CH964239 KRF99636.1 CH954180 EDV46357.1 AAAB01008847 EAA45046.5 M28231 AF050085 AF050084 AE014298 AY058284 BT024971 X76243 X76244 KZ288194 PBC33965.1 EDW82547.1 ACZ95239.1 ACZ95241.1 AGB95180.1 KQS29945.1 CM000162 EDX02240.1 KRK06659.1 CH379066 KRT07328.1 ACZ95240.1 AGB95181.1 EAL31379.2 KRT07327.1 CH902640 KPU77357.1 KRK06660.1 OUUW01000003 SPP78106.1 SPP78105.1 EDV38314.2 KPU77356.1 GFTR01008644 JAW07782.1 GFDL01007336 JAV27709.1 ATLV01017216 KE525157 KFB42062.1 GFDL01007361 JAV27684.1 CH479235 EDW36693.1 UFQS01002439 UFQT01002439 SSX14044.1 SSX33463.1 CH933810 KRF94266.1 GGFM01000594 MBW21345.1 EGK96582.1 KQ414667 KOC64766.1 GBXI01000988 JAD13304.1 GGFJ01000760 MBW49901.1 EDW07901.2 KRF94267.1 GAMC01019325 GAMC01019324 GAMC01019323 GAMC01019322 JAB87233.1 ADMH02001982 ETN60086.1 KQ434809 KZC06285.1 CH916371 EDV92457.1 KQS29947.1 KQ435863 KOX70321.1 AFH07295.1 GGFM01000603 MBW21354.1 GGFM01000599 MBW21350.1 GGFK01001442 MBW34763.1 CH940651 EDW65583.2 KRK06658.1 GGFJ01000748 MBW49889.1 ADTU01008311 ADTU01008312 GDHF01030716 GDHF01014633 GDHF01004025 JAI21598.1 JAI37681.1 JAI48289.1 GDHF01029359 GDHF01024218 GDHF01005058 JAI22955.1 JAI28096.1 JAI47256.1 GGFJ01000761 MBW49902.1 GGFK01001463 MBW34784.1 GAKP01020419 GAKP01020418 JAC38534.1 SPP78107.1 GGFM01000602 MBW21353.1 CH480850 EDW51334.1 CP012528 ALC49261.1 CCAG010021144 JXJN01005350 AJVK01026363 AJVK01026364 AJVK01026365 AJVK01026366 AJVK01026367 KQ982174 KYQ59371.1 CH477503 EAT39893.1 AAZX01014469 KRF94268.1 UFQT01002340 SSX33234.1
U50719 AAC47451.1 AGBW02013246 OWR43836.1 KQ459597 KPI95162.1 KQ460970 KPJ10193.1 GECU01002740 JAT04967.1 GFDF01006522 JAV07562.1 NEVH01024950 PNF16340.1 GANO01002435 JAB57436.1 PNF16341.1 PNF16342.1 DS235818 EEB17669.1 GBHO01002530 GBRD01018117 GDHC01017353 GDHC01003765 JAG41074.1 JAG47710.1 JAQ01276.1 JAQ14864.1 GBHO01015288 GBRD01017682 GDHC01016665 JAG28316.1 JAG48145.1 JAQ01964.1 JRES01000755 KNC28615.1 GFDL01007303 JAV27742.1 GFDL01007330 JAV27715.1 GFDL01007275 JAV27770.1 GFDL01007295 JAV27750.1 GFDL01007323 JAV27722.1 GBHO01015279 JAG28325.1 GECL01001809 JAP04315.1 GBHO01002526 JAG41078.1 CH964239 KRF99636.1 CH954180 EDV46357.1 AAAB01008847 EAA45046.5 M28231 AF050085 AF050084 AE014298 AY058284 BT024971 X76243 X76244 KZ288194 PBC33965.1 EDW82547.1 ACZ95239.1 ACZ95241.1 AGB95180.1 KQS29945.1 CM000162 EDX02240.1 KRK06659.1 CH379066 KRT07328.1 ACZ95240.1 AGB95181.1 EAL31379.2 KRT07327.1 CH902640 KPU77357.1 KRK06660.1 OUUW01000003 SPP78106.1 SPP78105.1 EDV38314.2 KPU77356.1 GFTR01008644 JAW07782.1 GFDL01007336 JAV27709.1 ATLV01017216 KE525157 KFB42062.1 GFDL01007361 JAV27684.1 CH479235 EDW36693.1 UFQS01002439 UFQT01002439 SSX14044.1 SSX33463.1 CH933810 KRF94266.1 GGFM01000594 MBW21345.1 EGK96582.1 KQ414667 KOC64766.1 GBXI01000988 JAD13304.1 GGFJ01000760 MBW49901.1 EDW07901.2 KRF94267.1 GAMC01019325 GAMC01019324 GAMC01019323 GAMC01019322 JAB87233.1 ADMH02001982 ETN60086.1 KQ434809 KZC06285.1 CH916371 EDV92457.1 KQS29947.1 KQ435863 KOX70321.1 AFH07295.1 GGFM01000603 MBW21354.1 GGFM01000599 MBW21350.1 GGFK01001442 MBW34763.1 CH940651 EDW65583.2 KRK06658.1 GGFJ01000748 MBW49889.1 ADTU01008311 ADTU01008312 GDHF01030716 GDHF01014633 GDHF01004025 JAI21598.1 JAI37681.1 JAI48289.1 GDHF01029359 GDHF01024218 GDHF01005058 JAI22955.1 JAI28096.1 JAI47256.1 GGFJ01000761 MBW49902.1 GGFK01001463 MBW34784.1 GAKP01020419 GAKP01020418 JAC38534.1 SPP78107.1 GGFM01000602 MBW21353.1 CH480850 EDW51334.1 CP012528 ALC49261.1 CCAG010021144 JXJN01005350 AJVK01026363 AJVK01026364 AJVK01026365 AJVK01026366 AJVK01026367 KQ982174 KYQ59371.1 CH477503 EAT39893.1 AAZX01014469 KRF94268.1 UFQT01002340 SSX33234.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000235965
UP000009046
UP000037069
+ More
UP000192221 UP000075903 UP000076407 UP000007798 UP000008711 UP000007062 UP000000803 UP000242457 UP000095301 UP000002282 UP000001819 UP000007801 UP000268350 UP000095300 UP000075884 UP000030765 UP000008744 UP000005203 UP000009192 UP000053825 UP000000673 UP000076502 UP000001070 UP000053105 UP000192223 UP000008792 UP000005205 UP000078200 UP000001292 UP000091820 UP000092553 UP000092444 UP000092460 UP000092462 UP000075809 UP000069272 UP000008820 UP000002358
UP000192221 UP000075903 UP000076407 UP000007798 UP000008711 UP000007062 UP000000803 UP000242457 UP000095301 UP000002282 UP000001819 UP000007801 UP000268350 UP000095300 UP000075884 UP000030765 UP000008744 UP000005203 UP000009192 UP000053825 UP000000673 UP000076502 UP000001070 UP000053105 UP000192223 UP000008792 UP000005205 UP000078200 UP000001292 UP000091820 UP000092553 UP000092444 UP000092460 UP000092462 UP000075809 UP000069272 UP000008820 UP000002358
Interpro
IPR003598
Ig_sub2
+ More
IPR036179 Ig-like_dom_sf
IPR026966 Neurofascin/L1/NrCAM_C
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR003529 Hematopoietin_rcpt_Gp130_CS
IPR013151 Immunoglobulin
IPR013106 Ig_V-set
IPR037604 Scm-like-4MBT1/2_SAM
IPR004092 Mbt
IPR021987 SLED
IPR038348 SLED_sf
IPR013761 SAM/pointed_sf
IPR036179 Ig-like_dom_sf
IPR026966 Neurofascin/L1/NrCAM_C
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR003529 Hematopoietin_rcpt_Gp130_CS
IPR013151 Immunoglobulin
IPR013106 Ig_V-set
IPR037604 Scm-like-4MBT1/2_SAM
IPR004092 Mbt
IPR021987 SLED
IPR038348 SLED_sf
IPR013761 SAM/pointed_sf
Gene 3D
ProteinModelPortal
D0VY44
A0A2W1BGE0
A0A2H1VMQ7
P91767
A0A212EQR2
A0A194PPC6
+ More
A0A0N1IE92 A0A1B6K0I8 A0A1L8DM81 A0A2J7PJ24 U5EU82 A0A2J7PJ23 A0A2J7PJ28 E0VWB3 A0A0A9Z9I6 A0A0A9Y9L8 A0A0L0C8N9 A0A1Q3FJR5 A0A1Q3FJL7 A0A1Q3FJH2 A0A1Q3FJF3 A0A1Q3FJP3 A0A0A9YFP0 A0A0V0G851 A0A0A9ZCK9 A0A1W4VUK4 A0A182V2N1 A0A182X573 A0A0Q9WUV5 B3NVG5 Q7PG72 P20241-2 A0A1W4VGI6 A0A2A3EQC2 B4NCI8 E1JJF9 A0A0Q5T3I4 A0A1I8M4Q6 B4Q0A3 A0A0R3P7J0 E1JJF8 P20241 Q29FM1 A0A0P8ZS96 A0A0R1EHH6 A0A3B0JXM4 A0A1I8PSB9 A0A3B0J8C5 B3N0B0 A0A1I8PS80 A0A182MZQ2 A0A224XGR9 A0A1Q3FJG2 A0A084VVR8 A0A1Q3FJI5 B4HA25 A0A088A0C7 A0A336L9G0 A0A0Q9X0E4 A0A2M3YYL2 F5HJW6 A0A0L7R1U9 A0A0A1XPS4 A0A2M4BA85 B4L315 W8ARL3 W5J7Y4 A0A154P2X8 B4JP14 A0A0Q5THL2 A0A0N1IT65 M9NEZ5 A0A2M3YYL5 A0A2M3YYM0 A0A2M4A1U8 A0A1W4X2R0 B4M1M9 A0A0R1EHL1 A0A2M4BA33 A0A158P3P7 A0A0K8WB81 A0A0K8W814 A0A2M4BA73 A0A2M4A1U2 A0A034V7N8 A0A3B0J7M4 A0A2M3YYJ9 A0A1A9V6L8 B4IJX8 A0A1A9WVD0 A0A0M5J3D9 A0A1B0G439 A0A1B0AXR8 A0A1B0GMP2 A0A151XGE0 A0A182FML2 Q16Z16 K7IUL4 A0A0Q9WVH7 A0A336MRR8
A0A0N1IE92 A0A1B6K0I8 A0A1L8DM81 A0A2J7PJ24 U5EU82 A0A2J7PJ23 A0A2J7PJ28 E0VWB3 A0A0A9Z9I6 A0A0A9Y9L8 A0A0L0C8N9 A0A1Q3FJR5 A0A1Q3FJL7 A0A1Q3FJH2 A0A1Q3FJF3 A0A1Q3FJP3 A0A0A9YFP0 A0A0V0G851 A0A0A9ZCK9 A0A1W4VUK4 A0A182V2N1 A0A182X573 A0A0Q9WUV5 B3NVG5 Q7PG72 P20241-2 A0A1W4VGI6 A0A2A3EQC2 B4NCI8 E1JJF9 A0A0Q5T3I4 A0A1I8M4Q6 B4Q0A3 A0A0R3P7J0 E1JJF8 P20241 Q29FM1 A0A0P8ZS96 A0A0R1EHH6 A0A3B0JXM4 A0A1I8PSB9 A0A3B0J8C5 B3N0B0 A0A1I8PS80 A0A182MZQ2 A0A224XGR9 A0A1Q3FJG2 A0A084VVR8 A0A1Q3FJI5 B4HA25 A0A088A0C7 A0A336L9G0 A0A0Q9X0E4 A0A2M3YYL2 F5HJW6 A0A0L7R1U9 A0A0A1XPS4 A0A2M4BA85 B4L315 W8ARL3 W5J7Y4 A0A154P2X8 B4JP14 A0A0Q5THL2 A0A0N1IT65 M9NEZ5 A0A2M3YYL5 A0A2M3YYM0 A0A2M4A1U8 A0A1W4X2R0 B4M1M9 A0A0R1EHL1 A0A2M4BA33 A0A158P3P7 A0A0K8WB81 A0A0K8W814 A0A2M4BA73 A0A2M4A1U2 A0A034V7N8 A0A3B0J7M4 A0A2M3YYJ9 A0A1A9V6L8 B4IJX8 A0A1A9WVD0 A0A0M5J3D9 A0A1B0G439 A0A1B0AXR8 A0A1B0GMP2 A0A151XGE0 A0A182FML2 Q16Z16 K7IUL4 A0A0Q9WVH7 A0A336MRR8
PDB
1CFB
E-value=6.38674e-64,
Score=624
Ontologies
GO
GO:0016021
GO:0004896
GO:0016319
GO:0098609
GO:0045924
GO:0072499
GO:0050808
GO:0048813
GO:0048675
GO:0007158
GO:0030175
GO:0008045
GO:0007560
GO:0048036
GO:0005918
GO:0005886
GO:0007409
GO:0019991
GO:0061343
GO:0005509
GO:0008050
GO:0021682
GO:0005923
GO:0035151
GO:0050839
GO:0005887
GO:0008049
GO:0008366
GO:0005919
GO:0060857
GO:0035011
GO:0006355
GO:0005634
GO:0042393
GO:0003714
GO:0005515
GO:0016020
GO:0004889
GO:0005216
GO:0006811
GO:0045211
GO:0006281
GO:0048384
Topology
Subcellular location
Cell membrane
Cell junction
Tight junction
Cell junction
Tight junction
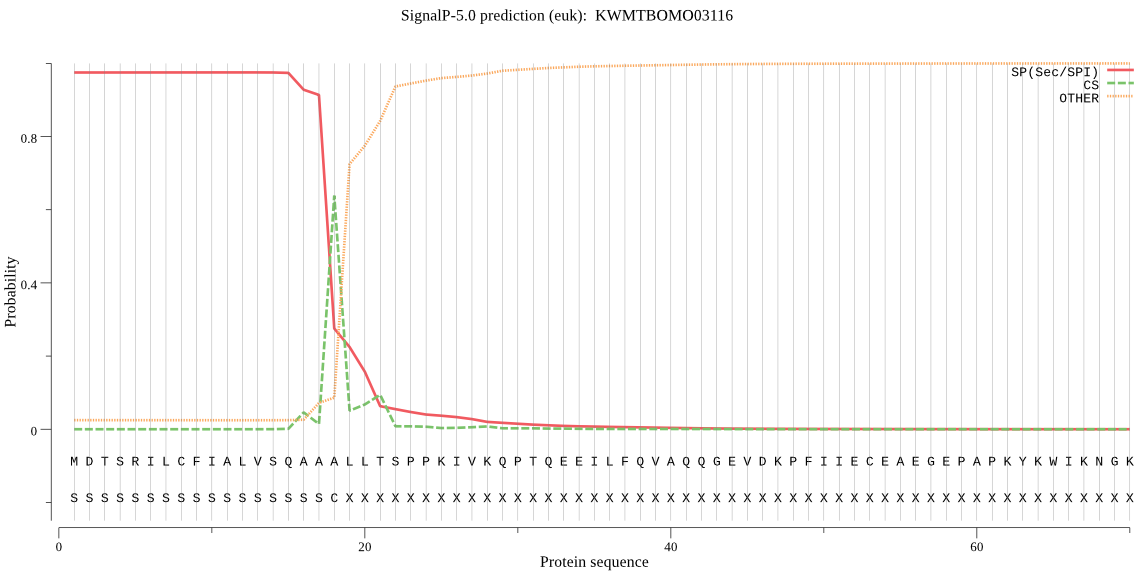
SignalP
Position: 1 - 18,
Likelihood: 0.975086
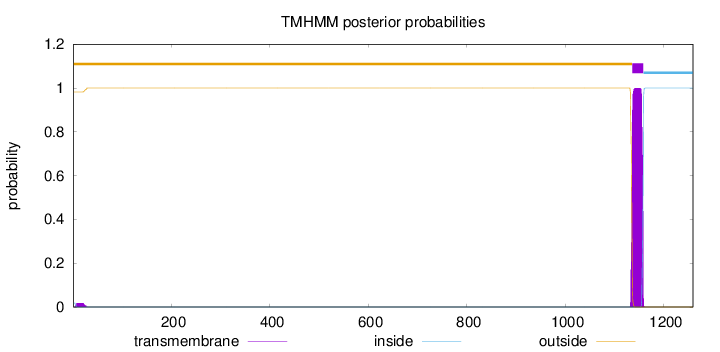
Length:
1259
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.20957
Exp number, first 60 AAs:
0.35459
Total prob of N-in:
0.01828
outside
1 - 1135
TMhelix
1136 - 1158
inside
1159 - 1259
Population Genetic Test Statistics
Pi
20.714032
Theta
19.793308
Tajima's D
0.007913
CLR
1.667313
CSRT
0.378831058447078
Interpretation
Uncertain
