Gene
KWMTBOMO03114
Pre Gene Modal
BGIBMGA005797
Annotation
PREDICTED:_CSC1-like_protein_2_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.961
Sequence
CDS
ATGAGTCGTTCTACCAACGAAATGTTATTCAGCCTATCGGACACCCCGAATGGGACGATCGATGATAGCTGCATGCTAGTGAACACATCGCAGAGTATTATCATCACGGGTGCCTACAATGGAATCCCTCAGACTTTGATCCTAAATTTGATAGCATGGTCGTTCCTTATTTTATTGTTTACTGTGCTTCGAAGGACGGCATGGAACTACGGGAGAATGGCACTCGTCCAGCGCACCAGGGGTCGCTGGACAAACTTATTCTATCGCGCTCGAGATGGTGATGCATTGGTCGAGCGCCGCCGCAGTGCAGACGAGTCTTCAAACATTGACGAAGGGCTCTTTACGTGGGTGCCGGCCGTGTTTCGCATCACTCGTGAACAGGTTCTCATAAAGTGTGGTCCTGATGCTGTGCACTATCTTTCCTTCCAGAGACACGCTATTACACTAATGTTCATCATAACCTTAGCATCCATTGGTATTATCTTGCCCATAAATTTCCAAGGGACCAATTTAAATGTGGACACTGGAACATTTAGTCGAACAACACTTTCCAACTTAAATGGAAGATCTTACTGGCTTTGGGTACATACCTTACTGTGCATCTCTTTCTTACCTATATCAGTGCTTATCATGAGAAGATGCACTGGGCGCAAACCCGTCCCTACATCTATGACTGGGACCATCATGATAACGAACATAGCCAAGGCTGACAGAAATGAGTCAACCATTCGAGCCTATTTTGCTCACAACTTTCCTCATATACAAATTGTAGACCTTCGCTTAGCTTATAACATCAACAAGCTCAATGAAGTTTTAGAACAAAAAGAAATGGTAACTGAAGCTTTAATCTACACTGATACATATTTCAAGAAGACTGGGAAACGTATTGCTGTTAAACCAACTTTGTGTGGACCTGAAATTGATGCTTTGGAATTTTACACAAATGAAGAAAAGAGGTTAAAAGATGAAGCTAAGAGAAGTCGTGCCATGGTCCTCAATGATCCTTTGGGAGTTGCCTTCTTAACTTTACCCTCATATCAGCTAGCTGAACATGTGATGAACCACTTCAGTCTCCATCGTGGCTGGGTGCTGTCACATGCTACTAACCCTGCAGATATTATTTGGGAGAATCTAAGTGTGAAGCCTGGAATCTGGTATGTTAAGGCCATCATAGTCAACTTTTTCCTCTTTGTTGTTTTGTTTTTCTTGACTACTCCTGCATTTGTTGTTAGTTTGTTCAACAGGACCGTGGTCAAACCAGATTCTCCTAACAAAATTACTTCTATCATCTATGATTTCTTGCCCACATTATTATTATGGACCATGGCTGCAGTGATGCCAGCCATTGTAGCATTCTCTGATAAATTCTTGTCACACTGGACCAAGTCACAGCAAAATTATTCAATTATGACAAAAACTGTATCATATCTCCTACTGATGACCTTGATCTTGCCATCATTAGGTCTCGCCAGTGCCGAGGCCTTCCTTATGTGGACATTACAACACCAGAATAACACAATGCGTTGGGACTGCGTGTTTCTCCCTGACAAGGGTGCTTTCTTTGTGAATTATGTTATCACCTCAGGCTTTATTGGTACTTCATTAGAGTTGATTCGTTTTCCGGAATTGTTCTTATATGTGTGGTATCTATTGCAATCAAAATCGAGAGCGGAAAAAAGTTACGTGAAAAAGGCTATATTGTACGAATTCCCATTCGGCGTGCACTACGCTTGGAGCATTGCGATATTTTCCATCACCACGGTGTACAGTCTGGCGTGCCCGCTGATCGCGCCGTTCGGGCTCATCTATCTAATGCTGAAGCACGTGGGGGACAAGCACAACTTGTACTTCGCGTACGGGCCAAGCGACATGAGCGGCATGGGCGGTGGTCGGATCCACGCCACCGCTGTCAGACTCATACGAGTCTCCGTTCTCCTCTTACTGATCAGCATGGCCGCCTGGGCTGGTCTCCGGGCCGGCTTCGAAGCTAGAACTATAATATTGATAATGGCCTCGATTGTGGCTTTCGGGACGTTTTTGCTGTTGAGTCCGTTTCCGAGTTGCACCCCACCCGCGCCCCTGCAGGCCGAGTCCAGTGTGAGGTTCCAAGAGTACATCGCCCCCGTGCTACTAAAGCCGGTGGATACGCCCCCGACAACGACGCCGACGACCCCGGATTACGGGGCATCATCGCCTGTTAACAACAATTCATACAATTCGGGCCCCATCAACATTTAA
Protein
MSRSTNEMLFSLSDTPNGTIDDSCMLVNTSQSIIITGAYNGIPQTLILNLIAWSFLILLFTVLRRTAWNYGRMALVQRTRGRWTNLFYRARDGDALVERRRSADESSNIDEGLFTWVPAVFRITREQVLIKCGPDAVHYLSFQRHAITLMFIITLASIGIILPINFQGTNLNVDTGTFSRTTLSNLNGRSYWLWVHTLLCISFLPISVLIMRRCTGRKPVPTSMTGTIMITNIAKADRNESTIRAYFAHNFPHIQIVDLRLAYNINKLNEVLEQKEMVTEALIYTDTYFKKTGKRIAVKPTLCGPEIDALEFYTNEEKRLKDEAKRSRAMVLNDPLGVAFLTLPSYQLAEHVMNHFSLHRGWVLSHATNPADIIWENLSVKPGIWYVKAIIVNFFLFVVLFFLTTPAFVVSLFNRTVVKPDSPNKITSIIYDFLPTLLLWTMAAVMPAIVAFSDKFLSHWTKSQQNYSIMTKTVSYLLLMTLILPSLGLASAEAFLMWTLQHQNNTMRWDCVFLPDKGAFFVNYVITSGFIGTSLELIRFPELFLYVWYLLQSKSRAEKSYVKKAILYEFPFGVHYAWSIAIFSITTVYSLACPLIAPFGLIYLMLKHVGDKHNLYFAYGPSDMSGMGGGRIHATAVRLIRVSVLLLLISMAAWAGLRAGFEARTIILIMASIVAFGTFLLLSPFPSCTPPAPLQAESSVRFQEYIAPVLLKPVDTPPTTTPTTPDYGASSPVNNNSYNSGPINI
Summary
Uniprot
A0A2A4JBN6
A0A2W1BBW0
A0A194PQB6
A0A0N0PBD1
A0A2H1VNU0
A0A0L7LNI9
+ More
Q16T32 B0WVM3 A0A0K8TLU9 A0A1L8DDM3 A0A1Q3FAH4 A0A1Q3FD11 A0A084W7Q7 A0A182IWP7 A0A182RLB7 A0A182W3N5 A0A182KZC8 Q7Q7M1 A0A182V4P5 A0A182TU48 A0A182MLB9 A0A182PX35 A0A182Q8X9 A0A1Y1N6N8 A0A182NMZ4 A0A182T8V5 A0A182Y8J9 A0A2J7PMF8 A0A182FAK7 A0A1W4XJM8 A0A2M4BF71 A0A067R2P6 A0A2M4CHU3 A0A2M4AII8 W5JIB8 A0A2M4CHT9 A0A2M3Z675 B4NMJ5 N6T2H6 V5FZ93 A0A1I8QEA5 A0A0M4EAW1 A0A1A9WGL3 T1PFX9 Q6NP91 B3N988 A0A1Q3FAK4 B4LJM0 A0A182GX22 A0A0Q9W4P5 B4QFA5 A0A0J9R7S3 B4HRJ2 D6W8C0 B4J841 B4KNX3 B3MGE2 A0A3B0JVY7 A0A0A1WN72 A0A0T6B4R5 A0A2P8XK19 A0A1B0A3A0 A0A1A9VY27 A0A1B0G0A3 B4GGP0 W8B0H2 Q28ZW6 A0A1W4WB20 A0A1A9Y4S3 A0A1W4VZB5 A0A034W0K0 A0A0K8V6Z0 A0A0K8UQF7 A0A0K8UVY6 A0A1B0BE30 A0A336MRD6 A0A1B6DVT6 A0A0P4ZG53 A0A0P5FIL8 A0A0P5T2X0 A0A0P5JQ38 E9GMG1 A0A0N8ABI3 A0A0P5STS0 A0A0P5XHD6 A0A0P5UCG8 A0A0P5S9R1 A0A0P5DUX6 A0A0P6A1C7 A0A0P5GIX6 A0A0P5YP15 A0A0N7ZS72 A0A0P5DHB3 A0A0P4ZNV1 A0A0N8A1D3 A0A0M9A2K0 A0A0P5CGJ7 A0A0P5SX12 A0A0P5G1K8
Q16T32 B0WVM3 A0A0K8TLU9 A0A1L8DDM3 A0A1Q3FAH4 A0A1Q3FD11 A0A084W7Q7 A0A182IWP7 A0A182RLB7 A0A182W3N5 A0A182KZC8 Q7Q7M1 A0A182V4P5 A0A182TU48 A0A182MLB9 A0A182PX35 A0A182Q8X9 A0A1Y1N6N8 A0A182NMZ4 A0A182T8V5 A0A182Y8J9 A0A2J7PMF8 A0A182FAK7 A0A1W4XJM8 A0A2M4BF71 A0A067R2P6 A0A2M4CHU3 A0A2M4AII8 W5JIB8 A0A2M4CHT9 A0A2M3Z675 B4NMJ5 N6T2H6 V5FZ93 A0A1I8QEA5 A0A0M4EAW1 A0A1A9WGL3 T1PFX9 Q6NP91 B3N988 A0A1Q3FAK4 B4LJM0 A0A182GX22 A0A0Q9W4P5 B4QFA5 A0A0J9R7S3 B4HRJ2 D6W8C0 B4J841 B4KNX3 B3MGE2 A0A3B0JVY7 A0A0A1WN72 A0A0T6B4R5 A0A2P8XK19 A0A1B0A3A0 A0A1A9VY27 A0A1B0G0A3 B4GGP0 W8B0H2 Q28ZW6 A0A1W4WB20 A0A1A9Y4S3 A0A1W4VZB5 A0A034W0K0 A0A0K8V6Z0 A0A0K8UQF7 A0A0K8UVY6 A0A1B0BE30 A0A336MRD6 A0A1B6DVT6 A0A0P4ZG53 A0A0P5FIL8 A0A0P5T2X0 A0A0P5JQ38 E9GMG1 A0A0N8ABI3 A0A0P5STS0 A0A0P5XHD6 A0A0P5UCG8 A0A0P5S9R1 A0A0P5DUX6 A0A0P6A1C7 A0A0P5GIX6 A0A0P5YP15 A0A0N7ZS72 A0A0P5DHB3 A0A0P4ZNV1 A0A0N8A1D3 A0A0M9A2K0 A0A0P5CGJ7 A0A0P5SX12 A0A0P5G1K8
Pubmed
28756777
26354079
26227816
17510324
26369729
24438588
+ More
20966253 12364791 14747013 17210077 28004739 25244985 24845553 20920257 23761445 17994087 23537049 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 22936249 18362917 19820115 18057021 25830018 29403074 24495485 15632085 25348373 21292972
20966253 12364791 14747013 17210077 28004739 25244985 24845553 20920257 23761445 17994087 23537049 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 22936249 18362917 19820115 18057021 25830018 29403074 24495485 15632085 25348373 21292972
EMBL
NWSH01002243
PCG68842.1
KZ150156
PZC72779.1
KQ459597
KPI95158.1
+ More
KQ460970 KPJ10198.1 ODYU01003405 SOQ42122.1 JTDY01000494 KOB76924.1 CH477661 EAT37598.1 DS232128 EDS35630.1 GDAI01002251 JAI15352.1 GFDF01009532 JAV04552.1 GFDL01010461 JAV24584.1 GFDL01009646 JAV25399.1 ATLV01021279 ATLV01021280 ATLV01021281 KE525316 KFB46251.1 AAAB01008952 EAA10576.4 AXCM01007048 AXCN02002205 GEZM01014982 JAV91946.1 NEVH01023995 PNF17524.1 GGFJ01002460 MBW51601.1 KK852954 KDR13287.1 GGFL01000729 MBW64907.1 GGFK01007275 MBW40596.1 ADMH02001410 ETN62640.1 GGFL01000728 MBW64906.1 GGFM01003197 MBW23948.1 CH964282 EDW85584.1 APGK01047286 KB741077 KB632432 ENN74334.1 ERL95530.1 GALX01005456 JAB63010.1 CP012524 ALC40565.1 KA647050 AFP61679.1 AE013599 BT011035 AAF59136.1 AAR30195.1 CH954177 EDV59575.2 GFDL01010439 JAV24606.1 CH940648 EDW61588.2 JXUM01094518 JXUM01094519 KQ564134 KXJ72715.1 KRF80089.1 CM000362 EDX06134.1 CM002911 KMY92187.1 CH480816 EDW46874.1 KQ971307 EFA10885.1 CH916367 EDW02271.1 CH933808 EDW09018.1 KRG04425.1 KRG04426.1 CH902619 EDV37845.1 OUUW01000001 SPP75238.1 GBXI01014454 GBXI01005616 JAC99837.1 JAD08676.1 LJIG01009804 KRT82348.1 PYGN01001892 PSN32304.1 CCAG010021794 CH479183 EDW35660.1 GAMC01011959 GAMC01011957 GAMC01011955 GAMC01011953 JAB94600.1 CM000071 EAL25496.2 GAKP01009832 GAKP01009831 JAC49121.1 GDHF01017663 GDHF01016699 JAI34651.1 JAI35615.1 GDHF01023591 GDHF01011571 JAI28723.1 JAI40743.1 GDHF01021616 GDHF01018875 GDHF01006594 JAI30698.1 JAI33439.1 JAI45720.1 JXJN01012763 UFQT01002092 SSX32640.1 GEDC01007541 JAS29757.1 GDIP01213871 JAJ09531.1 GDIQ01254632 JAJ97092.1 GDIP01134130 JAL69584.1 GDIQ01200145 JAK51580.1 GL732552 EFX79398.1 GDIP01163673 JAJ59729.1 GDIP01135380 JAL68334.1 GDIP01072337 JAM31378.1 GDIP01115037 JAL88677.1 GDIQ01099181 JAL52545.1 GDIP01155076 JAJ68326.1 GDIP01040392 JAM63323.1 GDIQ01240488 JAK11237.1 GDIP01193921 GDIP01055146 JAM48569.1 GDIP01217761 JAJ05641.1 GDIP01157528 JAJ65874.1 GDIP01213870 JAJ09532.1 GDIP01192073 JAJ31329.1 KQ435783 KOX74643.1 GDIP01174568 JAJ48834.1 GDIP01134129 JAL69585.1 GDIQ01254631 JAJ97093.1
KQ460970 KPJ10198.1 ODYU01003405 SOQ42122.1 JTDY01000494 KOB76924.1 CH477661 EAT37598.1 DS232128 EDS35630.1 GDAI01002251 JAI15352.1 GFDF01009532 JAV04552.1 GFDL01010461 JAV24584.1 GFDL01009646 JAV25399.1 ATLV01021279 ATLV01021280 ATLV01021281 KE525316 KFB46251.1 AAAB01008952 EAA10576.4 AXCM01007048 AXCN02002205 GEZM01014982 JAV91946.1 NEVH01023995 PNF17524.1 GGFJ01002460 MBW51601.1 KK852954 KDR13287.1 GGFL01000729 MBW64907.1 GGFK01007275 MBW40596.1 ADMH02001410 ETN62640.1 GGFL01000728 MBW64906.1 GGFM01003197 MBW23948.1 CH964282 EDW85584.1 APGK01047286 KB741077 KB632432 ENN74334.1 ERL95530.1 GALX01005456 JAB63010.1 CP012524 ALC40565.1 KA647050 AFP61679.1 AE013599 BT011035 AAF59136.1 AAR30195.1 CH954177 EDV59575.2 GFDL01010439 JAV24606.1 CH940648 EDW61588.2 JXUM01094518 JXUM01094519 KQ564134 KXJ72715.1 KRF80089.1 CM000362 EDX06134.1 CM002911 KMY92187.1 CH480816 EDW46874.1 KQ971307 EFA10885.1 CH916367 EDW02271.1 CH933808 EDW09018.1 KRG04425.1 KRG04426.1 CH902619 EDV37845.1 OUUW01000001 SPP75238.1 GBXI01014454 GBXI01005616 JAC99837.1 JAD08676.1 LJIG01009804 KRT82348.1 PYGN01001892 PSN32304.1 CCAG010021794 CH479183 EDW35660.1 GAMC01011959 GAMC01011957 GAMC01011955 GAMC01011953 JAB94600.1 CM000071 EAL25496.2 GAKP01009832 GAKP01009831 JAC49121.1 GDHF01017663 GDHF01016699 JAI34651.1 JAI35615.1 GDHF01023591 GDHF01011571 JAI28723.1 JAI40743.1 GDHF01021616 GDHF01018875 GDHF01006594 JAI30698.1 JAI33439.1 JAI45720.1 JXJN01012763 UFQT01002092 SSX32640.1 GEDC01007541 JAS29757.1 GDIP01213871 JAJ09531.1 GDIQ01254632 JAJ97092.1 GDIP01134130 JAL69584.1 GDIQ01200145 JAK51580.1 GL732552 EFX79398.1 GDIP01163673 JAJ59729.1 GDIP01135380 JAL68334.1 GDIP01072337 JAM31378.1 GDIP01115037 JAL88677.1 GDIQ01099181 JAL52545.1 GDIP01155076 JAJ68326.1 GDIP01040392 JAM63323.1 GDIQ01240488 JAK11237.1 GDIP01193921 GDIP01055146 JAM48569.1 GDIP01217761 JAJ05641.1 GDIP01157528 JAJ65874.1 GDIP01213870 JAJ09532.1 GDIP01192073 JAJ31329.1 KQ435783 KOX74643.1 GDIP01174568 JAJ48834.1 GDIP01134129 JAL69585.1 GDIQ01254631 JAJ97093.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000037510
UP000008820
UP000002320
+ More
UP000030765 UP000075880 UP000075900 UP000075920 UP000075882 UP000007062 UP000075903 UP000075902 UP000075883 UP000075885 UP000075886 UP000075884 UP000075901 UP000076408 UP000235965 UP000069272 UP000192223 UP000027135 UP000000673 UP000007798 UP000019118 UP000030742 UP000095300 UP000092553 UP000091820 UP000095301 UP000000803 UP000008711 UP000008792 UP000069940 UP000249989 UP000000304 UP000001292 UP000007266 UP000001070 UP000009192 UP000007801 UP000268350 UP000245037 UP000092445 UP000078200 UP000092444 UP000008744 UP000001819 UP000192221 UP000092443 UP000092460 UP000000305 UP000053105
UP000030765 UP000075880 UP000075900 UP000075920 UP000075882 UP000007062 UP000075903 UP000075902 UP000075883 UP000075885 UP000075886 UP000075884 UP000075901 UP000076408 UP000235965 UP000069272 UP000192223 UP000027135 UP000000673 UP000007798 UP000019118 UP000030742 UP000095300 UP000092553 UP000091820 UP000095301 UP000000803 UP000008711 UP000008792 UP000069940 UP000249989 UP000000304 UP000001292 UP000007266 UP000001070 UP000009192 UP000007801 UP000268350 UP000245037 UP000092445 UP000078200 UP000092444 UP000008744 UP000001819 UP000192221 UP000092443 UP000092460 UP000000305 UP000053105
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4JBN6
A0A2W1BBW0
A0A194PQB6
A0A0N0PBD1
A0A2H1VNU0
A0A0L7LNI9
+ More
Q16T32 B0WVM3 A0A0K8TLU9 A0A1L8DDM3 A0A1Q3FAH4 A0A1Q3FD11 A0A084W7Q7 A0A182IWP7 A0A182RLB7 A0A182W3N5 A0A182KZC8 Q7Q7M1 A0A182V4P5 A0A182TU48 A0A182MLB9 A0A182PX35 A0A182Q8X9 A0A1Y1N6N8 A0A182NMZ4 A0A182T8V5 A0A182Y8J9 A0A2J7PMF8 A0A182FAK7 A0A1W4XJM8 A0A2M4BF71 A0A067R2P6 A0A2M4CHU3 A0A2M4AII8 W5JIB8 A0A2M4CHT9 A0A2M3Z675 B4NMJ5 N6T2H6 V5FZ93 A0A1I8QEA5 A0A0M4EAW1 A0A1A9WGL3 T1PFX9 Q6NP91 B3N988 A0A1Q3FAK4 B4LJM0 A0A182GX22 A0A0Q9W4P5 B4QFA5 A0A0J9R7S3 B4HRJ2 D6W8C0 B4J841 B4KNX3 B3MGE2 A0A3B0JVY7 A0A0A1WN72 A0A0T6B4R5 A0A2P8XK19 A0A1B0A3A0 A0A1A9VY27 A0A1B0G0A3 B4GGP0 W8B0H2 Q28ZW6 A0A1W4WB20 A0A1A9Y4S3 A0A1W4VZB5 A0A034W0K0 A0A0K8V6Z0 A0A0K8UQF7 A0A0K8UVY6 A0A1B0BE30 A0A336MRD6 A0A1B6DVT6 A0A0P4ZG53 A0A0P5FIL8 A0A0P5T2X0 A0A0P5JQ38 E9GMG1 A0A0N8ABI3 A0A0P5STS0 A0A0P5XHD6 A0A0P5UCG8 A0A0P5S9R1 A0A0P5DUX6 A0A0P6A1C7 A0A0P5GIX6 A0A0P5YP15 A0A0N7ZS72 A0A0P5DHB3 A0A0P4ZNV1 A0A0N8A1D3 A0A0M9A2K0 A0A0P5CGJ7 A0A0P5SX12 A0A0P5G1K8
Q16T32 B0WVM3 A0A0K8TLU9 A0A1L8DDM3 A0A1Q3FAH4 A0A1Q3FD11 A0A084W7Q7 A0A182IWP7 A0A182RLB7 A0A182W3N5 A0A182KZC8 Q7Q7M1 A0A182V4P5 A0A182TU48 A0A182MLB9 A0A182PX35 A0A182Q8X9 A0A1Y1N6N8 A0A182NMZ4 A0A182T8V5 A0A182Y8J9 A0A2J7PMF8 A0A182FAK7 A0A1W4XJM8 A0A2M4BF71 A0A067R2P6 A0A2M4CHU3 A0A2M4AII8 W5JIB8 A0A2M4CHT9 A0A2M3Z675 B4NMJ5 N6T2H6 V5FZ93 A0A1I8QEA5 A0A0M4EAW1 A0A1A9WGL3 T1PFX9 Q6NP91 B3N988 A0A1Q3FAK4 B4LJM0 A0A182GX22 A0A0Q9W4P5 B4QFA5 A0A0J9R7S3 B4HRJ2 D6W8C0 B4J841 B4KNX3 B3MGE2 A0A3B0JVY7 A0A0A1WN72 A0A0T6B4R5 A0A2P8XK19 A0A1B0A3A0 A0A1A9VY27 A0A1B0G0A3 B4GGP0 W8B0H2 Q28ZW6 A0A1W4WB20 A0A1A9Y4S3 A0A1W4VZB5 A0A034W0K0 A0A0K8V6Z0 A0A0K8UQF7 A0A0K8UVY6 A0A1B0BE30 A0A336MRD6 A0A1B6DVT6 A0A0P4ZG53 A0A0P5FIL8 A0A0P5T2X0 A0A0P5JQ38 E9GMG1 A0A0N8ABI3 A0A0P5STS0 A0A0P5XHD6 A0A0P5UCG8 A0A0P5S9R1 A0A0P5DUX6 A0A0P6A1C7 A0A0P5GIX6 A0A0P5YP15 A0A0N7ZS72 A0A0P5DHB3 A0A0P4ZNV1 A0A0N8A1D3 A0A0M9A2K0 A0A0P5CGJ7 A0A0P5SX12 A0A0P5G1K8
PDB
6MGW
E-value=2.12302e-19,
Score=238
Ontologies
GO
PANTHER
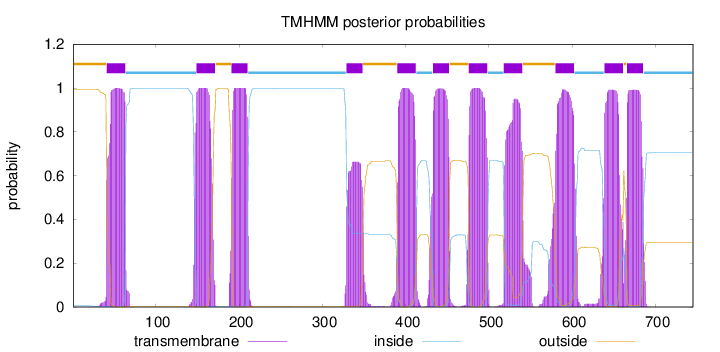
Topology
Length:
745
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
235.36551
Exp number, first 60 AAs:
18.69927
Total prob of N-in:
0.00525
POSSIBLE N-term signal
sequence
outside
1 - 40
TMhelix
41 - 63
inside
64 - 148
TMhelix
149 - 171
outside
172 - 190
TMhelix
191 - 210
inside
211 - 328
TMhelix
329 - 348
outside
349 - 389
TMhelix
390 - 412
inside
413 - 432
TMhelix
433 - 452
outside
453 - 475
TMhelix
476 - 498
inside
499 - 517
TMhelix
518 - 540
outside
541 - 579
TMhelix
580 - 602
inside
603 - 638
TMhelix
639 - 661
outside
662 - 665
TMhelix
666 - 685
inside
686 - 745
Population Genetic Test Statistics
Pi
6.321079
Theta
6.794573
Tajima's D
0.040725
CLR
0.900735
CSRT
0.397230138493075
Interpretation
Uncertain
