Gene
KWMTBOMO03113 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005769
Annotation
signal_sequence_receptor_beta_subunit_precursor_[Bombyx_mori]
Full name
Translocon-associated protein subunit beta
Alternative Name
Signal sequence receptor subunit beta
Location in the cell
Extracellular Reliability : 1.008 PlasmaMembrane Reliability : 1.429
Sequence
CDS
ATGGCAGCGAAACTGTTATATTTTGTACTCTTCATCGCTGCCGCAGTGTCTGCGGATGATGAACCCGTGCTAGCAAGACTACTAGTGTCAAAACAAGTGCTAAACAAATATTTAGTAGAGAATATGGATATACTAGTAAAATACACATTGTTCAATGTTGGGAGCGCACCGGCTGTTGAAGTGAAACTCGTGGACAACGGCTTCCATCCGGACGTGTTTACTGTAGTAGGCGGACAACTGACCGCGGAAATCGATAGGATTGCACCTCAAACTAATGTATCCCATGTAGTGACTGTCCGGTCCAATAAGTACGGCTATTTCAACTTCAGCTCGGCGGAAGTCACATACAAAGCCAGCGAAGACGCCACAGACGTCCAATATTCGATCAGCAGTGCTCCAGGAGAGGGAGCCATCGTAGCCTTCAAAGATTATGATAGAAAGTTCTCCTCGCACATCCTCGATTGGGCGGCATTCGCAGTTATGACCCTCCCGTCCCTAGCCATTCCATTTGGCCTTTGGTACTCATCAAAGAGCAAATATGAGAAGCTTTCAAAACCCAAAAAGGGACACTAG
Protein
MAAKLLYFVLFIAAAVSADDEPVLARLLVSKQVLNKYLVENMDILVKYTLFNVGSAPAVEVKLVDNGFHPDVFTVVGGQLTAEIDRIAPQTNVSHVVTVRSNKYGYFNFSSAEVTYKASEDATDVQYSISSAPGEGAIVAFKDYDRKFSSHILDWAAFAVMTLPSLAIPFGLWYSSKSKYEKLSKPKKGH
Summary
Description
TRAP proteins are part of a complex whose function is to bind calcium to the ER membrane and thereby regulate the retention of ER resident proteins.
Subunit
Heterotetramer of TRAP-alpha, TRAP-beta, TRAP-delta and TRAP-gamma.
Similarity
Belongs to the TRAP-beta family.
Feature
chain Translocon-associated protein subunit beta
Uniprot
Q2F5V9
I4DIY9
A0A1E1W4W0
A0A0N1ICH5
A0A2W1BJ75
A0A2A4JBR7
+ More
A0A2H1VMR9 I4DMV9 A0A0L7LNX3 S4PTH9 A0A212EP32 H9J8C7 A0A0T6AUQ0 A0A336L0A9 A0A1Y1LW79 T1DF53 A0A2J7RKQ5 A0A023EIX1 Q1HR42 A0A023EIH6 A0A2P8XPC6 U5EUQ5 A0A182U1Q1 A0A182XMC1 Q5TRB4 A0A182I256 A0A1B6L274 A0A182KPY5 A0A182PTN0 A0A182FH18 A0A182MDI9 A0A182V9I5 A0A1B6EIC8 A0A1B6IFF5 T1E7Z5 A0A182T7E7 A0A2M4A2Y6 A0A2M3Z035 A0A2M3Z040 A0A182J9D2 W5J5L7 A0A2M4C0E0 A0A182WLC2 A0A182R4I6 A0A182YEF2 A0A182NJD5 A0A182JRR4 B0WEU7 A0A182PZQ2 A0A1Q3FBQ0 A0A310SFR8 A0A2A3EUL7 A0A088AJ89 A0A154PM41 D6W8B9 A0A084WEB2 A0A026W5F5 A0A151HYM0 A0A0L7RBV4 E0VR09 A0A1B0DNI9 A0A195ESC3 F4X4S3 E9J4Q8 A0A0M5JAY6 A0A3B0JY80 A0A151II83 A0A1L8DV92 B4H293 Q29EN1 B4IU56 Q6XIK3 A0A1B6CTP5 A0A1W4UUL6 B4HJ22 B3NI52 B4QLU5 B4IZY6 B4MM43 B3M957 Q9VUZ0 B4L9P5 A0A1I8MQN4 B4LDE1 A0A0J7NXD8 A0A0Q9XQW9 A0A0A9YUM2 C4N188 A0A0L0CLW7 E2AX30 A0A1I8P1G1 E2BC60 A2I433 A0A232FKP8 E9GL54 A0A0P5L9K2 A0A1A9YH05 A0A1B0BC61 A0A0P5G2H6 A0A1A9ZKF8
A0A2H1VMR9 I4DMV9 A0A0L7LNX3 S4PTH9 A0A212EP32 H9J8C7 A0A0T6AUQ0 A0A336L0A9 A0A1Y1LW79 T1DF53 A0A2J7RKQ5 A0A023EIX1 Q1HR42 A0A023EIH6 A0A2P8XPC6 U5EUQ5 A0A182U1Q1 A0A182XMC1 Q5TRB4 A0A182I256 A0A1B6L274 A0A182KPY5 A0A182PTN0 A0A182FH18 A0A182MDI9 A0A182V9I5 A0A1B6EIC8 A0A1B6IFF5 T1E7Z5 A0A182T7E7 A0A2M4A2Y6 A0A2M3Z035 A0A2M3Z040 A0A182J9D2 W5J5L7 A0A2M4C0E0 A0A182WLC2 A0A182R4I6 A0A182YEF2 A0A182NJD5 A0A182JRR4 B0WEU7 A0A182PZQ2 A0A1Q3FBQ0 A0A310SFR8 A0A2A3EUL7 A0A088AJ89 A0A154PM41 D6W8B9 A0A084WEB2 A0A026W5F5 A0A151HYM0 A0A0L7RBV4 E0VR09 A0A1B0DNI9 A0A195ESC3 F4X4S3 E9J4Q8 A0A0M5JAY6 A0A3B0JY80 A0A151II83 A0A1L8DV92 B4H293 Q29EN1 B4IU56 Q6XIK3 A0A1B6CTP5 A0A1W4UUL6 B4HJ22 B3NI52 B4QLU5 B4IZY6 B4MM43 B3M957 Q9VUZ0 B4L9P5 A0A1I8MQN4 B4LDE1 A0A0J7NXD8 A0A0Q9XQW9 A0A0A9YUM2 C4N188 A0A0L0CLW7 E2AX30 A0A1I8P1G1 E2BC60 A2I433 A0A232FKP8 E9GL54 A0A0P5L9K2 A0A1A9YH05 A0A1B0BC61 A0A0P5G2H6 A0A1A9ZKF8
Pubmed
22651552
26354079
28756777
26227816
23622113
22118469
+ More
19121390 28004739 24330624 24945155 17204158 17510324 26483478 29403074 12364791 14747013 17210077 20966253 20920257 23761445 25244985 18362917 19820115 24438588 24508170 30249741 20566863 21719571 21282665 17994087 15632085 17550304 14525923 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25401762 26823975 19576987 26108605 20798317 28648823 21292972
19121390 28004739 24330624 24945155 17204158 17510324 26483478 29403074 12364791 14747013 17210077 20966253 20920257 23761445 25244985 18362917 19820115 24438588 24508170 30249741 20566863 21719571 21282665 17994087 15632085 17550304 14525923 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25401762 26823975 19576987 26108605 20798317 28648823 21292972
EMBL
DQ311314
ABD36258.1
AK401257
KQ459597
BAM17879.1
KPI95157.1
+ More
GDQN01009143 GDQN01007697 JAT81911.1 JAT83357.1 KQ460970 KPJ10199.1 KZ150156 PZC72780.1 NWSH01002243 PCG68843.1 ODYU01003405 SOQ42121.1 AK402627 BAM19249.1 JTDY01000494 KOB76921.1 GAIX01012008 JAA80552.1 AGBW02013553 OWR43217.1 BABH01027343 LJIG01022755 KRT78895.1 UFQS01001397 UFQT01001397 SSX10685.1 SSX30367.1 GEZM01048396 JAV76590.1 GALA01000782 JAA94070.1 NEVH01002717 PNF41412.1 GAPW01004687 JAC08911.1 DQ440252 CH477632 ABF18285.1 EAT38041.1 JXUM01003522 GAPW01004686 GAPW01004685 KQ560159 JAC08912.1 KXJ84118.1 PYGN01001589 PSN33858.1 GANO01002218 JAB57653.1 AAAB01008960 EAL39950.2 APCN01000122 GEBQ01022179 JAT17798.1 AXCM01002346 AXCM01017416 GECZ01032128 JAS37641.1 GECU01022079 JAS85627.1 GAMD01003203 JAA98387.1 GGFK01001794 MBW35115.1 GGFM01001100 MBW21851.1 GGFM01001134 MBW21885.1 ADMH02002052 ETN59747.1 GGFJ01009635 MBW58776.1 DS231911 EDS45860.1 AXCN02000448 GFDL01010041 JAV25004.1 KQ768636 OAD53089.1 KZ288185 PBC34972.1 KQ434977 KZC12827.1 KQ971307 EFA10929.1 ATLV01023198 KE525341 KFB48556.1 KK107405 QOIP01000014 EZA51233.1 RLU14974.1 KQ976730 KYM76456.1 KQ414617 KOC68236.1 DS235442 EEB15815.1 AJVK01017579 KQ981993 KYN31071.1 GL888679 EGI58496.1 GL768112 EFZ12206.1 CP012525 ALC43181.1 OUUW01000012 SPP87004.1 KQ977505 KYN02270.1 GFDF01003751 JAV10333.1 CH479203 EDW30445.1 CH379070 EAL30029.1 CM000159 CH891789 EDW95481.1 EDW99919.1 AY231827 AAR09850.1 GEDC01020446 JAS16852.1 CH480815 EDW41678.1 CH954178 EDV52208.1 CM000363 CM002912 EDX10644.1 KMY99952.1 CH916366 EDV96758.1 CH963847 EDW73052.1 KRF97684.1 CH902618 EDV40041.1 AF160923 AE014296 AAD46863.1 AAF49532.2 CH933816 EDW17093.1 CH940647 EDW68879.1 LBMM01001019 KMQ97060.1 KRG07604.1 GBHO01006912 GBRD01014381 GDHC01010993 JAG36692.1 JAG51445.1 JAQ07636.1 EZ048861 ACN69153.1 JRES01000215 KNC33232.1 GL443520 EFN61980.1 GL447255 EFN86736.1 EF070536 ABM55602.1 NNAY01000081 OXU31163.1 GL732550 EFX79793.1 GDIQ01174529 JAK77196.1 JXJN01011858 GDIQ01247260 JAK04465.1
GDQN01009143 GDQN01007697 JAT81911.1 JAT83357.1 KQ460970 KPJ10199.1 KZ150156 PZC72780.1 NWSH01002243 PCG68843.1 ODYU01003405 SOQ42121.1 AK402627 BAM19249.1 JTDY01000494 KOB76921.1 GAIX01012008 JAA80552.1 AGBW02013553 OWR43217.1 BABH01027343 LJIG01022755 KRT78895.1 UFQS01001397 UFQT01001397 SSX10685.1 SSX30367.1 GEZM01048396 JAV76590.1 GALA01000782 JAA94070.1 NEVH01002717 PNF41412.1 GAPW01004687 JAC08911.1 DQ440252 CH477632 ABF18285.1 EAT38041.1 JXUM01003522 GAPW01004686 GAPW01004685 KQ560159 JAC08912.1 KXJ84118.1 PYGN01001589 PSN33858.1 GANO01002218 JAB57653.1 AAAB01008960 EAL39950.2 APCN01000122 GEBQ01022179 JAT17798.1 AXCM01002346 AXCM01017416 GECZ01032128 JAS37641.1 GECU01022079 JAS85627.1 GAMD01003203 JAA98387.1 GGFK01001794 MBW35115.1 GGFM01001100 MBW21851.1 GGFM01001134 MBW21885.1 ADMH02002052 ETN59747.1 GGFJ01009635 MBW58776.1 DS231911 EDS45860.1 AXCN02000448 GFDL01010041 JAV25004.1 KQ768636 OAD53089.1 KZ288185 PBC34972.1 KQ434977 KZC12827.1 KQ971307 EFA10929.1 ATLV01023198 KE525341 KFB48556.1 KK107405 QOIP01000014 EZA51233.1 RLU14974.1 KQ976730 KYM76456.1 KQ414617 KOC68236.1 DS235442 EEB15815.1 AJVK01017579 KQ981993 KYN31071.1 GL888679 EGI58496.1 GL768112 EFZ12206.1 CP012525 ALC43181.1 OUUW01000012 SPP87004.1 KQ977505 KYN02270.1 GFDF01003751 JAV10333.1 CH479203 EDW30445.1 CH379070 EAL30029.1 CM000159 CH891789 EDW95481.1 EDW99919.1 AY231827 AAR09850.1 GEDC01020446 JAS16852.1 CH480815 EDW41678.1 CH954178 EDV52208.1 CM000363 CM002912 EDX10644.1 KMY99952.1 CH916366 EDV96758.1 CH963847 EDW73052.1 KRF97684.1 CH902618 EDV40041.1 AF160923 AE014296 AAD46863.1 AAF49532.2 CH933816 EDW17093.1 CH940647 EDW68879.1 LBMM01001019 KMQ97060.1 KRG07604.1 GBHO01006912 GBRD01014381 GDHC01010993 JAG36692.1 JAG51445.1 JAQ07636.1 EZ048861 ACN69153.1 JRES01000215 KNC33232.1 GL443520 EFN61980.1 GL447255 EFN86736.1 EF070536 ABM55602.1 NNAY01000081 OXU31163.1 GL732550 EFX79793.1 GDIQ01174529 JAK77196.1 JXJN01011858 GDIQ01247260 JAK04465.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000037510
UP000007151
UP000005204
+ More
UP000235965 UP000008820 UP000069940 UP000249989 UP000245037 UP000075902 UP000076407 UP000007062 UP000075840 UP000075882 UP000075885 UP000069272 UP000075883 UP000075903 UP000075901 UP000075880 UP000000673 UP000075920 UP000075900 UP000076408 UP000075884 UP000075881 UP000002320 UP000075886 UP000242457 UP000005203 UP000076502 UP000007266 UP000030765 UP000053097 UP000279307 UP000078540 UP000053825 UP000009046 UP000092462 UP000078541 UP000007755 UP000092553 UP000268350 UP000078542 UP000008744 UP000001819 UP000002282 UP000192221 UP000001292 UP000008711 UP000000304 UP000001070 UP000007798 UP000007801 UP000000803 UP000009192 UP000095301 UP000008792 UP000036403 UP000037069 UP000000311 UP000095300 UP000008237 UP000215335 UP000000305 UP000092443 UP000092460 UP000092445
UP000235965 UP000008820 UP000069940 UP000249989 UP000245037 UP000075902 UP000076407 UP000007062 UP000075840 UP000075882 UP000075885 UP000069272 UP000075883 UP000075903 UP000075901 UP000075880 UP000000673 UP000075920 UP000075900 UP000076408 UP000075884 UP000075881 UP000002320 UP000075886 UP000242457 UP000005203 UP000076502 UP000007266 UP000030765 UP000053097 UP000279307 UP000078540 UP000053825 UP000009046 UP000092462 UP000078541 UP000007755 UP000092553 UP000268350 UP000078542 UP000008744 UP000001819 UP000002282 UP000192221 UP000001292 UP000008711 UP000000304 UP000001070 UP000007798 UP000007801 UP000000803 UP000009192 UP000095301 UP000008792 UP000036403 UP000037069 UP000000311 UP000095300 UP000008237 UP000215335 UP000000305 UP000092443 UP000092460 UP000092445
PRIDE
Interpro
IPR008856
TRAP_beta
ProteinModelPortal
Q2F5V9
I4DIY9
A0A1E1W4W0
A0A0N1ICH5
A0A2W1BJ75
A0A2A4JBR7
+ More
A0A2H1VMR9 I4DMV9 A0A0L7LNX3 S4PTH9 A0A212EP32 H9J8C7 A0A0T6AUQ0 A0A336L0A9 A0A1Y1LW79 T1DF53 A0A2J7RKQ5 A0A023EIX1 Q1HR42 A0A023EIH6 A0A2P8XPC6 U5EUQ5 A0A182U1Q1 A0A182XMC1 Q5TRB4 A0A182I256 A0A1B6L274 A0A182KPY5 A0A182PTN0 A0A182FH18 A0A182MDI9 A0A182V9I5 A0A1B6EIC8 A0A1B6IFF5 T1E7Z5 A0A182T7E7 A0A2M4A2Y6 A0A2M3Z035 A0A2M3Z040 A0A182J9D2 W5J5L7 A0A2M4C0E0 A0A182WLC2 A0A182R4I6 A0A182YEF2 A0A182NJD5 A0A182JRR4 B0WEU7 A0A182PZQ2 A0A1Q3FBQ0 A0A310SFR8 A0A2A3EUL7 A0A088AJ89 A0A154PM41 D6W8B9 A0A084WEB2 A0A026W5F5 A0A151HYM0 A0A0L7RBV4 E0VR09 A0A1B0DNI9 A0A195ESC3 F4X4S3 E9J4Q8 A0A0M5JAY6 A0A3B0JY80 A0A151II83 A0A1L8DV92 B4H293 Q29EN1 B4IU56 Q6XIK3 A0A1B6CTP5 A0A1W4UUL6 B4HJ22 B3NI52 B4QLU5 B4IZY6 B4MM43 B3M957 Q9VUZ0 B4L9P5 A0A1I8MQN4 B4LDE1 A0A0J7NXD8 A0A0Q9XQW9 A0A0A9YUM2 C4N188 A0A0L0CLW7 E2AX30 A0A1I8P1G1 E2BC60 A2I433 A0A232FKP8 E9GL54 A0A0P5L9K2 A0A1A9YH05 A0A1B0BC61 A0A0P5G2H6 A0A1A9ZKF8
A0A2H1VMR9 I4DMV9 A0A0L7LNX3 S4PTH9 A0A212EP32 H9J8C7 A0A0T6AUQ0 A0A336L0A9 A0A1Y1LW79 T1DF53 A0A2J7RKQ5 A0A023EIX1 Q1HR42 A0A023EIH6 A0A2P8XPC6 U5EUQ5 A0A182U1Q1 A0A182XMC1 Q5TRB4 A0A182I256 A0A1B6L274 A0A182KPY5 A0A182PTN0 A0A182FH18 A0A182MDI9 A0A182V9I5 A0A1B6EIC8 A0A1B6IFF5 T1E7Z5 A0A182T7E7 A0A2M4A2Y6 A0A2M3Z035 A0A2M3Z040 A0A182J9D2 W5J5L7 A0A2M4C0E0 A0A182WLC2 A0A182R4I6 A0A182YEF2 A0A182NJD5 A0A182JRR4 B0WEU7 A0A182PZQ2 A0A1Q3FBQ0 A0A310SFR8 A0A2A3EUL7 A0A088AJ89 A0A154PM41 D6W8B9 A0A084WEB2 A0A026W5F5 A0A151HYM0 A0A0L7RBV4 E0VR09 A0A1B0DNI9 A0A195ESC3 F4X4S3 E9J4Q8 A0A0M5JAY6 A0A3B0JY80 A0A151II83 A0A1L8DV92 B4H293 Q29EN1 B4IU56 Q6XIK3 A0A1B6CTP5 A0A1W4UUL6 B4HJ22 B3NI52 B4QLU5 B4IZY6 B4MM43 B3M957 Q9VUZ0 B4L9P5 A0A1I8MQN4 B4LDE1 A0A0J7NXD8 A0A0Q9XQW9 A0A0A9YUM2 C4N188 A0A0L0CLW7 E2AX30 A0A1I8P1G1 E2BC60 A2I433 A0A232FKP8 E9GL54 A0A0P5L9K2 A0A1A9YH05 A0A1B0BC61 A0A0P5G2H6 A0A1A9ZKF8
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
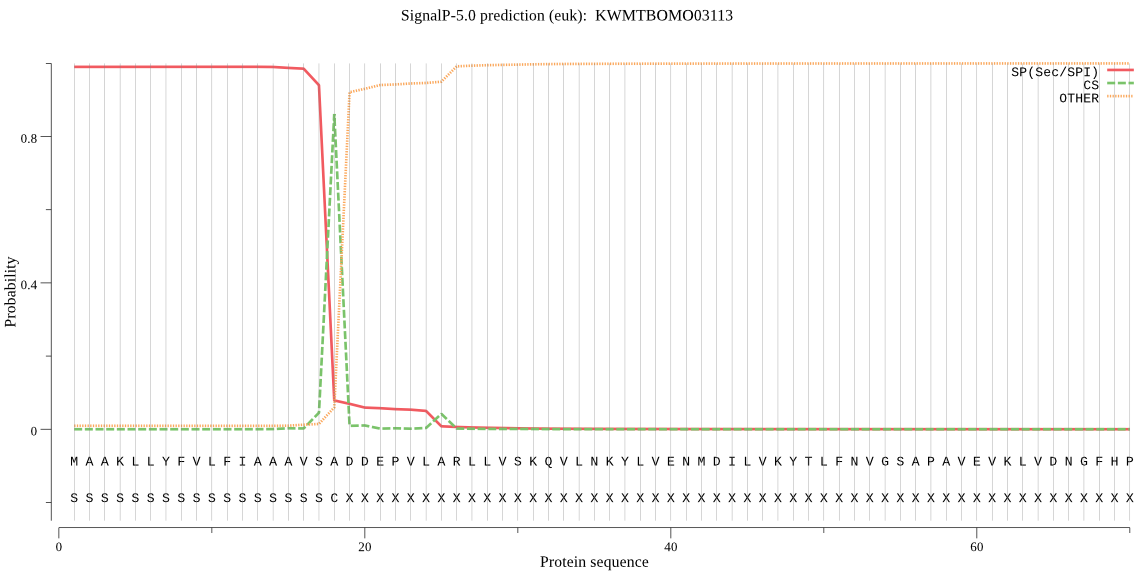
SignalP
Position: 1 - 18,
Likelihood: 0.990115
Length:
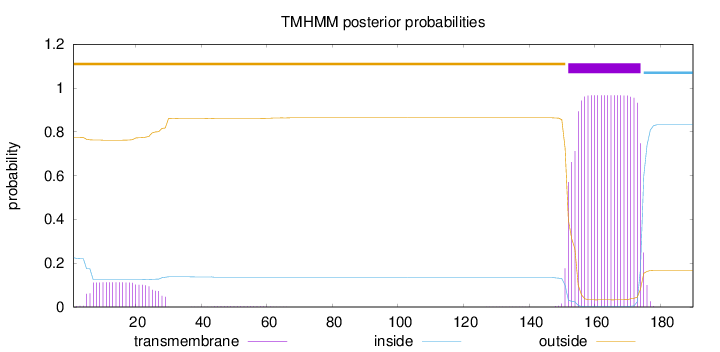
190
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.94817
Exp number, first 60 AAs:
2.46927
Total prob of N-in:
0.22547
outside
1 - 151
TMhelix
152 - 174
inside
175 - 190
Population Genetic Test Statistics
Pi
16.676175
Theta
20.229826
Tajima's D
-0.539307
CLR
0
CSRT
0.233188340582971
Interpretation
Uncertain
