Pre Gene Modal
BGIBMGA005798
Annotation
PREDICTED:_centromere/kinetochore_protein_zw10_homolog_[Amyelois_transitella]
Full name
Centromere/kinetochore protein zw10 homolog
Location in the cell
Cytoplasmic Reliability : 3.494
Sequence
CDS
ATGACAGATAAAGAAGATGTCGACAATAAACCTGAAAATGTTCCTATAGATGACGAAGCCCTAGAGAAGTTGGCTGTTGAAGAACTGATACGTGAAGCTGTTCAAGGCGCCGCTAGAGCTGAAATCGTTGGTCCGTCAGGTTGGGTTAAAGCCCCTCCTCAAAAAACCAACAAACGTTTTCTGATAAGTCAAAAAGAAAACGTTAAAATGGAGAGTTATGATATAGAAGAAGTTTTAAAAAATATATTTGATTGTAATGAAACAAACTGGAAACCAGCTGACAAAATACCCCTTCTAACCAAAGAAATAGAACTGTTAAAAATAGATTTAAAATCTTATTTGGAAACTTGTGAGTATAATGTAGATTCAGTTTTGGAGGAGAGTAAAATATTGTTTGTGGATAGTAAAGCCGTTGTTGATGAACTTGAAAAATGCAAGAAGGAAATAGAAGAGCACACAATGGCAGAAGTGCTCAAATCCATCGAAACCCATGACCAGATCACTAAAGAGTTGCAGCATGTTAATTTTGCACTGAACATTGTGTATGATGTTGGACAGTGTGGTAAACTTGTGAAGCAATTTGAAGAAGGAAGACAGGCACAAAGTTACACACGTGCTGTGGAGGCTGTGTATGATTTACTCCAATATATAGAGCTACCTGCTGAAGGTTTCAAATATTTAGATATATATGCTAACACAAAACATACAGCTCAACTTATACTGGATAAACTCCTGCAAGACCTGTATACAGAATGGACAAGACTTGTGTCATGGAATACTAAAGTCAGCTCAAAGAAATCAGTAGTTAAAATTACTTTGAAAATTGAAGATACACCATTTGTTATCGATGTGATAAGAGCATTGGATAAAAATAAAAATTTGAAAGAAAAAGTTAGTGAGTTCTCTCAGTTCTTGTATAAAGAAGTTCTACAGCCAATCATTTATAACGACTGTATAGTTTTCCCAGAAACTGAAGAGCTATTGACAGTAACAATAAATTACAAGCAAAATTATAAGCCTCCTTATGATCAAGTTATAGCCAACTTTAGGTTACTTTTCCATCACTTGAGTTCCAAATTGAAATTTGAATATGATAAGAATTGTAGTATTATGAAGTTGATTGGGAATGAAATTAGCGGACCGTTCAGTGAGCTGATCATGAAAGACTGTTTCATAGACACAATCCCAAATAATATTATAGATCTACAAACATATGGTAAAATTACTTCAGAGATAGAAAAATTTCAGCATTTCCTTTTCATTGTGAAGTTCTTTACAGAAGATATATCTATATTAAAATACATAGAAGATATAGATATCTTATTCTCAAAGAAATCTAGTGATCACTTCTTGAAGACAGCTAGAGACATAATGCTGAAAGACCTAAGTGTTTGTATGTCTATTGGTGTTGAGAATATTTCTGATGAAGAAGAATCAAGTGATGATTATGGAAAGGCAGCTGAAGCCTTGAGAATACTTGAAAAGACAATACCAAAGAGTTTATTCTACTTTCCGAGGTGTATGATATCAAAAACAGCTCAAGAGCTTCTAGATTTGATTTATGTCATAATGGAGCAAGCTGTGCAGTGTTCTGATGTAGTTTGTAAGAAATTATATCAGACCACCAGGTTAATATTTGAGCTCTATGATGCAGTGATGCCATATCACCATGAAAAATACTTGCAGACTATTCCTCAGTATGTTGCATTATTCCACAACAACTGTATGTATCTTGCACACAATATGATGACCTTTGGTGATAAGTGGTTAACATTGATGGAGGGTCGAGACATCGATTATCCCATTGGCTTTGTTGATTTGGTTCAGAAGTTACGGGAACTGGGTTACAAGCACCTTACAATACATCTGCAGCAGCAGAGGAAACAAATCTTGGATAACATACGTTCTTCAGATCTCAACTGCATGGTGGTCAAAGAAGTGTTAGGCTCCAATGCTGAGGCCGCTATCCGCCAGTGCTTGCGGCAACTACAGCTTTTGAAAAATGTGTGGATAGGAGTGTTCCCTATCAATGTCTTCACAAGGTTGATGGCGACCCTAGTCAACATGTTCATCGAGGAACTCATTCACCGAGTGTGCACCGTCGAGGACATCTCAATGGAGATGGCGACACAGCTGACCGAGATATATACATTAGTAGTGCAAAAGGCACCACAATTGTTCCAGAGTTCAACAGATATAGAAACTCACGTCAAATCCTGGATAAAGCTGCAGGAGCTAATCTTTGTCCTTGGAGGGTCTCTGAAGGATATCGACAACCACTGGAAGGATGGTACCGGTCCCTTGGCGGTCCACTTCAGAACCGAAGAGTTACGGAATCTAATTAAGGCTCTCTTCCAGAATACTCAGATTCGAGCCAACTTGCTGTCTAAAATAAAGTAA
Protein
MTDKEDVDNKPENVPIDDEALEKLAVEELIREAVQGAARAEIVGPSGWVKAPPQKTNKRFLISQKENVKMESYDIEEVLKNIFDCNETNWKPADKIPLLTKEIELLKIDLKSYLETCEYNVDSVLEESKILFVDSKAVVDELEKCKKEIEEHTMAEVLKSIETHDQITKELQHVNFALNIVYDVGQCGKLVKQFEEGRQAQSYTRAVEAVYDLLQYIELPAEGFKYLDIYANTKHTAQLILDKLLQDLYTEWTRLVSWNTKVSSKKSVVKITLKIEDTPFVIDVIRALDKNKNLKEKVSEFSQFLYKEVLQPIIYNDCIVFPETEELLTVTINYKQNYKPPYDQVIANFRLLFHHLSSKLKFEYDKNCSIMKLIGNEISGPFSELIMKDCFIDTIPNNIIDLQTYGKITSEIEKFQHFLFIVKFFTEDISILKYIEDIDILFSKKSSDHFLKTARDIMLKDLSVCMSIGVENISDEEESSDDYGKAAEALRILEKTIPKSLFYFPRCMISKTAQELLDLIYVIMEQAVQCSDVVCKKLYQTTRLIFELYDAVMPYHHEKYLQTIPQYVALFHNNCMYLAHNMMTFGDKWLTLMEGRDIDYPIGFVDLVQKLRELGYKHLTIHLQQQRKQILDNIRSSDLNCMVVKEVLGSNAEAAIRQCLRQLQLLKNVWIGVFPINVFTRLMATLVNMFIEELIHRVCTVEDISMEMATQLTEIYTLVVQKAPQLFQSSTDIETHVKSWIKLQELIFVLGGSLKDIDNHWKDGTGPLAVHFRTEELRNLIKALFQNTQIRANLLSKIK
Summary
Description
Essential component of the mitotic checkpoint, which prevents cells from prematurely exiting mitosis. Required for the assembly of the dynein-dynactin and MAD1-MAD2 complexes onto kinetochores. Its function related to the spindle assembly machinery is proposed to depend on its association in the mitotic RZZ complex. Involved in regulation of membrane traffic between the Golgi and the endoplasmic reticulum (ER); the function is proposed to depend on its association in the interphase NRZ complex which is believed to play a role in SNARE assembly at the ER (By similarity).
Subunit
Interacts with NBAS and KNTC1/ROD; the interactions are mutually exclusive and indicative for its association in two different vesicle tethering complexes. Component of the RZZ complex composed of KNTC1/ROD, ZW10 and ZWILCH. Component of the NRZ complex composed of NBAS, ZW10 and RINT1/TIP20L; NRZ associates with SNAREs STX18, USE1L, BNIP1/SEC20L and SEC22B (the assembly has been described as syntaxin 18 complex). Interacts directly with RINT1/TIP20L bound to BNIP1/SEC20L. Interacts with C19orf25 and ZWINT STOPPED Associated with a SNARE complex consisting of STX18, USE1L, BNIP1/SEC20L, and SEC22B through direct interaction with RINT1/TIP20L bound to BNIP1/SEC20L. Component of the RZZ complex composed of KNTC1/ROD, ZW10 and ZWILCH. Interacts with C19orf25, KNTC1 and ZWINT.
Similarity
Belongs to the peptidase C19 family.
Belongs to the ZW10 family.
Belongs to the ZW10 family.
Keywords
Acetylation
Cell cycle
Cell division
Centromere
Chromosome
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Endoplasmic reticulum
ER-Golgi transport
Kinetochore
Membrane
Mitosis
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Centromere/kinetochore protein zw10 homolog
Uniprot
H9J8F6
A0A2W1BEN9
A0A2H1VMR4
A0A1E1WK00
A0A0N1PFC5
A0A212EP10
+ More
A0A194PVK0 A0A0L7LNX8 A0A2A4JBB9 A0A067R1H8 A0A2J7R4X7 A0A154PD87 A0A088ADK1 A0A2J7R4Y7 A0A232F9F2 K7IZY4 A0A3L8DW78 A0A0C9R7R5 A0A0L7RGC3 F4WFS7 A0A151JRX3 A0A158NYR6 E2C1W0 A0A026WMT4 A0A195CIN0 A0A1B6DLN4 E2AR01 A0A1B6K948 E9IYQ5 A0A1B6HJZ5 A0A195CW51 A0A195BTF8 A0A151X3F9 A0A0M8ZP20 E0VWF2 A0A093QFG8 K1PVH5 V4B230 V9KF16 A0A1L8DX26 A0A091R5P9 L5KMC6 R0M7L5 A0A091MXR0 A0A023EY20 A0A3Q0GZ42 A0A210QXB8 G3WIT0 A0A151P0I0 A0A093EZ14 M3W382 A0A0K8TM68 A0A093TJZ0 A0A091KDI5 A0A2I0LQY5 A0A093CLN2 U3IRS3 A0A091RB81 A0A087QSR9 A0A091TNP4 A0A384C086 A0A091WUP7 F6U6L7 A0A091M5E7 A0A0A0A176 A0A1W4XM69 A0A091J057 A0A0V0GA94 F6UZG3 A0A2T7NYV5 Q4V8C2 A0A091T0H9 M3XXR3 G3SN94 A0A069DZH5 B1H3L7 A0A1W2WAN0 A0A1V4J916 A0A0P5SCK6 H0YPQ8 A0A0P6I7F2 A0A091Q497 A0A1S3IFT2 A0A164QZG2 A0A094LHK6 H9GEQ3
A0A194PVK0 A0A0L7LNX8 A0A2A4JBB9 A0A067R1H8 A0A2J7R4X7 A0A154PD87 A0A088ADK1 A0A2J7R4Y7 A0A232F9F2 K7IZY4 A0A3L8DW78 A0A0C9R7R5 A0A0L7RGC3 F4WFS7 A0A151JRX3 A0A158NYR6 E2C1W0 A0A026WMT4 A0A195CIN0 A0A1B6DLN4 E2AR01 A0A1B6K948 E9IYQ5 A0A1B6HJZ5 A0A195CW51 A0A195BTF8 A0A151X3F9 A0A0M8ZP20 E0VWF2 A0A093QFG8 K1PVH5 V4B230 V9KF16 A0A1L8DX26 A0A091R5P9 L5KMC6 R0M7L5 A0A091MXR0 A0A023EY20 A0A3Q0GZ42 A0A210QXB8 G3WIT0 A0A151P0I0 A0A093EZ14 M3W382 A0A0K8TM68 A0A093TJZ0 A0A091KDI5 A0A2I0LQY5 A0A093CLN2 U3IRS3 A0A091RB81 A0A087QSR9 A0A091TNP4 A0A384C086 A0A091WUP7 F6U6L7 A0A091M5E7 A0A0A0A176 A0A1W4XM69 A0A091J057 A0A0V0GA94 F6UZG3 A0A2T7NYV5 Q4V8C2 A0A091T0H9 M3XXR3 G3SN94 A0A069DZH5 B1H3L7 A0A1W2WAN0 A0A1V4J916 A0A0P5SCK6 H0YPQ8 A0A0P6I7F2 A0A091Q497 A0A1S3IFT2 A0A164QZG2 A0A094LHK6 H9GEQ3
Pubmed
19121390
28756777
26354079
22118469
26227816
24845553
+ More
28648823 20075255 30249741 21719571 21347285 20798317 24508170 21282665 20566863 22992520 23254933 24402279 23258410 25474469 28812685 21709235 22293439 17975172 26369729 23371554 23749191 24813606 17495919 16341006 15489334 26334808 20360741 21881562
28648823 20075255 30249741 21719571 21347285 20798317 24508170 21282665 20566863 22992520 23254933 24402279 23258410 25474469 28812685 21709235 22293439 17975172 26369729 23371554 23749191 24813606 17495919 16341006 15489334 26334808 20360741 21881562
EMBL
BABH01027343
KZ150156
PZC72781.1
ODYU01003405
SOQ42119.1
GDQN01003721
+ More
JAT87333.1 KQ460970 KPJ10200.1 AGBW02013553 OWR43219.1 KQ459597 KPI95155.1 JTDY01000494 KOB76926.1 NWSH01002243 PCG68844.1 KK852777 KDR16765.1 NEVH01007399 PNF35887.1 KQ434875 KZC09767.1 PNF35889.1 NNAY01000597 OXU27491.1 QOIP01000003 RLU24714.1 GBYB01002786 JAG72553.1 KQ414596 KOC70027.1 GL888122 EGI66970.1 KQ978566 KYN30123.1 ADTU01004235 GL452008 EFN78072.1 KK107151 EZA57238.1 KQ977696 KYN00573.1 GEDC01010709 JAS26589.1 GL441846 EFN64147.1 GEBQ01032000 JAT07977.1 GL767043 EFZ14278.1 GECU01032755 JAS74951.1 KQ977220 KYN04906.1 KQ976407 KYM91386.1 KQ982566 KYQ54748.1 KQ435944 KOX68210.1 DS235819 EEB17703.1 KL672246 KFW85130.1 JH818606 EKC28357.1 KB200454 ESP01626.1 JW863892 AFO96409.1 GFDF01003074 JAV11010.1 KK713957 KFQ35143.1 KB030667 ELK11906.1 KB742431 EOB08693.1 KK838361 KFP81419.1 GBBI01004781 JAC13931.1 NEDP02001349 OWF53385.1 AEFK01114682 AEFK01114683 AEFK01114684 AKHW03001374 KYO42657.1 KK378720 KFV47216.1 AANG04000369 GDAI01002377 JAI15226.1 KL442499 KFW94564.1 KK552259 KFP34128.1 AKCR02000134 PKK19841.1 KL462244 KFV13869.1 ADON01004108 ADON01004109 KK813349 KFQ37113.1 KL225870 KFM04273.1 KK459094 KFQ78064.1 KL411261 KFR05171.1 KK521647 KFP66646.1 KL870337 KGL87717.1 KK501366 KFP14354.1 GECL01001267 JAP04857.1 AAEX03003483 AAEX03003484 PZQS01000008 PVD26313.1 BC097452 KK494687 KFQ64909.1 AEYP01026721 GBGD01000630 JAC88259.1 BC161442 AAI61442.1 LSYS01008642 OPJ68267.1 GDIQ01095484 JAL56242.1 ABQF01045786 ABQF01045787 ABQF01045788 GDIQ01009472 JAN85265.1 KK685443 KFQ14376.1 LRGB01002285 KZS08205.1 KL357880 KFZ63424.1 AAWZ02025845 AAWZ02025846 AAWZ02025847 AAWZ02025848
JAT87333.1 KQ460970 KPJ10200.1 AGBW02013553 OWR43219.1 KQ459597 KPI95155.1 JTDY01000494 KOB76926.1 NWSH01002243 PCG68844.1 KK852777 KDR16765.1 NEVH01007399 PNF35887.1 KQ434875 KZC09767.1 PNF35889.1 NNAY01000597 OXU27491.1 QOIP01000003 RLU24714.1 GBYB01002786 JAG72553.1 KQ414596 KOC70027.1 GL888122 EGI66970.1 KQ978566 KYN30123.1 ADTU01004235 GL452008 EFN78072.1 KK107151 EZA57238.1 KQ977696 KYN00573.1 GEDC01010709 JAS26589.1 GL441846 EFN64147.1 GEBQ01032000 JAT07977.1 GL767043 EFZ14278.1 GECU01032755 JAS74951.1 KQ977220 KYN04906.1 KQ976407 KYM91386.1 KQ982566 KYQ54748.1 KQ435944 KOX68210.1 DS235819 EEB17703.1 KL672246 KFW85130.1 JH818606 EKC28357.1 KB200454 ESP01626.1 JW863892 AFO96409.1 GFDF01003074 JAV11010.1 KK713957 KFQ35143.1 KB030667 ELK11906.1 KB742431 EOB08693.1 KK838361 KFP81419.1 GBBI01004781 JAC13931.1 NEDP02001349 OWF53385.1 AEFK01114682 AEFK01114683 AEFK01114684 AKHW03001374 KYO42657.1 KK378720 KFV47216.1 AANG04000369 GDAI01002377 JAI15226.1 KL442499 KFW94564.1 KK552259 KFP34128.1 AKCR02000134 PKK19841.1 KL462244 KFV13869.1 ADON01004108 ADON01004109 KK813349 KFQ37113.1 KL225870 KFM04273.1 KK459094 KFQ78064.1 KL411261 KFR05171.1 KK521647 KFP66646.1 KL870337 KGL87717.1 KK501366 KFP14354.1 GECL01001267 JAP04857.1 AAEX03003483 AAEX03003484 PZQS01000008 PVD26313.1 BC097452 KK494687 KFQ64909.1 AEYP01026721 GBGD01000630 JAC88259.1 BC161442 AAI61442.1 LSYS01008642 OPJ68267.1 GDIQ01095484 JAL56242.1 ABQF01045786 ABQF01045787 ABQF01045788 GDIQ01009472 JAN85265.1 KK685443 KFQ14376.1 LRGB01002285 KZS08205.1 KL357880 KFZ63424.1 AAWZ02025845 AAWZ02025846 AAWZ02025847 AAWZ02025848
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000037510
UP000218220
+ More
UP000027135 UP000235965 UP000076502 UP000005203 UP000215335 UP000002358 UP000279307 UP000053825 UP000007755 UP000078492 UP000005205 UP000008237 UP000053097 UP000078542 UP000000311 UP000078540 UP000075809 UP000053105 UP000009046 UP000053258 UP000005408 UP000030746 UP000010552 UP000189705 UP000242188 UP000007648 UP000050525 UP000011712 UP000053872 UP000016666 UP000053286 UP000261680 UP000291021 UP000053283 UP000002280 UP000053858 UP000192223 UP000053119 UP000002254 UP000245119 UP000002494 UP000000715 UP000007646 UP000190648 UP000007754 UP000085678 UP000076858 UP000001646
UP000027135 UP000235965 UP000076502 UP000005203 UP000215335 UP000002358 UP000279307 UP000053825 UP000007755 UP000078492 UP000005205 UP000008237 UP000053097 UP000078542 UP000000311 UP000078540 UP000075809 UP000053105 UP000009046 UP000053258 UP000005408 UP000030746 UP000010552 UP000189705 UP000242188 UP000007648 UP000050525 UP000011712 UP000053872 UP000016666 UP000053286 UP000261680 UP000291021 UP000053283 UP000002280 UP000053858 UP000192223 UP000053119 UP000002254 UP000245119 UP000002494 UP000000715 UP000007646 UP000190648 UP000007754 UP000085678 UP000076858 UP000001646
Interpro
Gene 3D
ProteinModelPortal
H9J8F6
A0A2W1BEN9
A0A2H1VMR4
A0A1E1WK00
A0A0N1PFC5
A0A212EP10
+ More
A0A194PVK0 A0A0L7LNX8 A0A2A4JBB9 A0A067R1H8 A0A2J7R4X7 A0A154PD87 A0A088ADK1 A0A2J7R4Y7 A0A232F9F2 K7IZY4 A0A3L8DW78 A0A0C9R7R5 A0A0L7RGC3 F4WFS7 A0A151JRX3 A0A158NYR6 E2C1W0 A0A026WMT4 A0A195CIN0 A0A1B6DLN4 E2AR01 A0A1B6K948 E9IYQ5 A0A1B6HJZ5 A0A195CW51 A0A195BTF8 A0A151X3F9 A0A0M8ZP20 E0VWF2 A0A093QFG8 K1PVH5 V4B230 V9KF16 A0A1L8DX26 A0A091R5P9 L5KMC6 R0M7L5 A0A091MXR0 A0A023EY20 A0A3Q0GZ42 A0A210QXB8 G3WIT0 A0A151P0I0 A0A093EZ14 M3W382 A0A0K8TM68 A0A093TJZ0 A0A091KDI5 A0A2I0LQY5 A0A093CLN2 U3IRS3 A0A091RB81 A0A087QSR9 A0A091TNP4 A0A384C086 A0A091WUP7 F6U6L7 A0A091M5E7 A0A0A0A176 A0A1W4XM69 A0A091J057 A0A0V0GA94 F6UZG3 A0A2T7NYV5 Q4V8C2 A0A091T0H9 M3XXR3 G3SN94 A0A069DZH5 B1H3L7 A0A1W2WAN0 A0A1V4J916 A0A0P5SCK6 H0YPQ8 A0A0P6I7F2 A0A091Q497 A0A1S3IFT2 A0A164QZG2 A0A094LHK6 H9GEQ3
A0A194PVK0 A0A0L7LNX8 A0A2A4JBB9 A0A067R1H8 A0A2J7R4X7 A0A154PD87 A0A088ADK1 A0A2J7R4Y7 A0A232F9F2 K7IZY4 A0A3L8DW78 A0A0C9R7R5 A0A0L7RGC3 F4WFS7 A0A151JRX3 A0A158NYR6 E2C1W0 A0A026WMT4 A0A195CIN0 A0A1B6DLN4 E2AR01 A0A1B6K948 E9IYQ5 A0A1B6HJZ5 A0A195CW51 A0A195BTF8 A0A151X3F9 A0A0M8ZP20 E0VWF2 A0A093QFG8 K1PVH5 V4B230 V9KF16 A0A1L8DX26 A0A091R5P9 L5KMC6 R0M7L5 A0A091MXR0 A0A023EY20 A0A3Q0GZ42 A0A210QXB8 G3WIT0 A0A151P0I0 A0A093EZ14 M3W382 A0A0K8TM68 A0A093TJZ0 A0A091KDI5 A0A2I0LQY5 A0A093CLN2 U3IRS3 A0A091RB81 A0A087QSR9 A0A091TNP4 A0A384C086 A0A091WUP7 F6U6L7 A0A091M5E7 A0A0A0A176 A0A1W4XM69 A0A091J057 A0A0V0GA94 F6UZG3 A0A2T7NYV5 Q4V8C2 A0A091T0H9 M3XXR3 G3SN94 A0A069DZH5 B1H3L7 A0A1W2WAN0 A0A1V4J916 A0A0P5SCK6 H0YPQ8 A0A0P6I7F2 A0A091Q497 A0A1S3IFT2 A0A164QZG2 A0A094LHK6 H9GEQ3
Ontologies
GO
GO:0000278
GO:0005634
GO:0000775
GO:0065003
GO:0006888
GO:1990423
GO:0005828
GO:0070939
GO:0007093
GO:0007080
GO:0000132
GO:0000922
GO:0034501
GO:0005829
GO:0007030
GO:0007096
GO:0007094
GO:0005737
GO:0016579
GO:0036459
GO:0006511
GO:0005783
GO:0051301
GO:0000777
GO:0000776
GO:0015031
GO:0000070
GO:0005789
GO:0007067
GO:0004889
GO:0005216
GO:0006811
GO:0016021
GO:0045211
GO:0006281
GO:0048384
Topology
Subcellular location
Cytoplasm
Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Endoplasmic reticulum membrane Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Chromosome Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Centromere Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Kinetochore Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Cytoskeleton Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Spindle Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Endoplasmic reticulum membrane Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Chromosome Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Centromere Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Kinetochore Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Cytoskeleton Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
Spindle Dynamic pattern of localization during the cell cycle. In most cells at interphase, present diffusely in the cytoplasm. In prometaphase, associated with the kinetochore. At metaphase, detected both at the kinetochores and, most prominently, at the spindle, particularly at the spindle poles. In very early anaphase, detected on segregating kinetochores. In late anaphase and telophase, accumulates at the spindle midzone. With evidence from 1 publications.
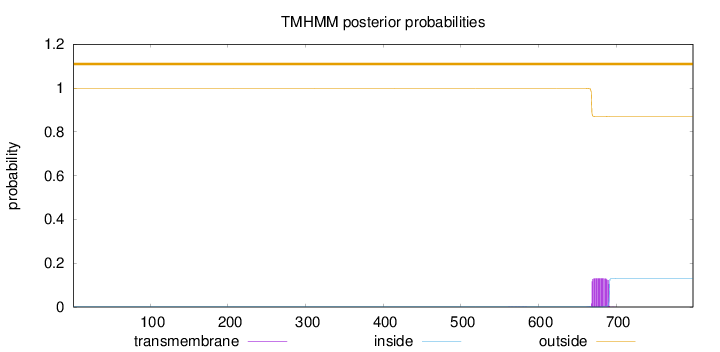
Length:
799
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.98119
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00184
outside
1 - 799
Population Genetic Test Statistics
Pi
17.69931
Theta
18.525685
Tajima's D
-0.703782
CLR
0.524532
CSRT
0.201389930503475
Interpretation
Uncertain
