Gene
KWMTBOMO03110
Pre Gene Modal
BGIBMGA005768
Annotation
hypothetical_protein_KGM_00604_[Danaus_plexippus]
Full name
Mediator of RNA polymerase II transcription subunit 27
Alternative Name
Mediator complex subunit 27
dCRSP34
dMED27
dTRAP37
dCRSP34
dMED27
dTRAP37
Location in the cell
Nuclear Reliability : 2.421
Sequence
CDS
ATGAATCAAACTATCAACGTAGAGCAATTAAATTCCGCTTTAAATTCACTACGGACACTACGATCGAGCGTCAGCCATGTATTCGAAACATTATCTAATGGACTACGCGCCGACCACGGCGAGGACGGCAAGGAGAAATTTCTATTAGAACTTCAAGAACTACTGAACAACGTTAATATAAACTTGAGGGACTTGGAACAGACAGTGAGTGGTCTCCCGATACCCACTGCTCCGCTGAATCTTGGTAATACCACGTATTTGAGTCAAGAAACCTCACAAGACAGGCAAACATTGTACACGCAGTTGGTTAACAGCTACAAATGGACTGATAAGATTCACGAATACAGCTCCTTCGCTCATACGTTACTGAGTCAGAACTCTCTCAAAAGGTCTTACATTAATTCTGGGAGTACGAAAAGAAGAGGCAAACTACAAAGTAATCACAATGTTGCTCCTATACAAGTTGATAATGTCATAAACAACATAGACAGATCCTATAACGATATGAAAATTACCATATCAAGACCATTTGCAAGTAATGCAGTTGTACAGATCAACCTAAGTCACGTTTTGAAAGCTGTTGTTGCTTTTAAGGGCTTATTGATGGAGTGGGTCATGGTGAAAGGCTATGGAGAGACCCTCGACCTGTGGACAGAGTCACGTCACCATGTATTTAGGAAAGTAACAGAAAATGCCCATGCAGCAATGTTACACTTCTACTCACCGACATTACCAGAGCTAGCAGTTAGGTCATTCATGACTTGGCTGCATAGTTATGTGAACCTTTTCAGTGAGCCATGTAAACGATGCAGTTGTCATTTACATCACACATCTCTGTTGCCACCAGCGTGGAGGGACTTCCGTACATTAGAAACATTTCATGATGAGTGCAAGCAATAG
Protein
MNQTINVEQLNSALNSLRTLRSSVSHVFETLSNGLRADHGEDGKEKFLLELQELLNNVNINLRDLEQTVSGLPIPTAPLNLGNTTYLSQETSQDRQTLYTQLVNSYKWTDKIHEYSSFAHTLLSQNSLKRSYINSGSTKRRGKLQSNHNVAPIQVDNVINNIDRSYNDMKITISRPFASNAVVQINLSHVLKAVVAFKGLLMEWVMVKGYGETLDLWTESRHHVFRKVTENAHAAMLHFYSPTLPELAVRSFMTWLHSYVNLFSEPCKRCSCHLHHTSLLPPAWRDFRTLETFHDECKQ
Summary
Description
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity).
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. Required for activated transcription of the MtnA gene.
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors. Required for activated transcription of the MtnA gene.
Subunit
Component of the Mediator complex.
Component of the Mediator complex, which includes at least CDK8, MED4, MED6, MED11, MED14, MED17, MED18, MED20, MED21, MED22, MED27, MED28, MED30 and MED31.
Component of the Mediator complex, which includes at least CDK8, MED4, MED6, MED11, MED14, MED17, MED18, MED20, MED21, MED22, MED27, MED28, MED30 and MED31.
Similarity
Belongs to the Mediator complex subunit 27 family.
Keywords
Activator
Complete proteome
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Mediator of RNA polymerase II transcription subunit 27
Uniprot
H9J8C6
A0A212EP38
A0A0L7LNA8
A0A0N1INM8
A0A2W1BM73
A0A2H1VMT2
+ More
A0A194PQB1 D2X8R5 A0A067R5Y6 A0A151IRJ9 A0A151IBA3 A0A195BTT5 A0A158NNR1 F4WV32 A0A195EW54 A0A151WSW2 A0A154PJQ4 A0A0M8ZVA6 A0A0L7R9J9 A0A026W257 A0A023F1U2 A0A224XUV3 A0A069DS88 E2C2M0 A0A2A3EJZ2 A0A087ZUD9 E0VTY9 A0A0C9RRE4 E2A3F7 A0A0A9WHB3 A0A0T6AYR4 A0A1Y1LMM7 A0A1B0CH03 A0A1B0DFY2 D6WH39 A0A1L8DFL3 K7J937 A0A182IS05 A0A232EZH6 A0A084W710 J3JUN3 A0A2M4AUD7 A0A182K701 W5JVP5 B0X296 A0A182P3V8 A0A182QAA0 A0A2J7QWU9 A0A182MLT5 A0A182VZY3 A0A2P8XYT8 A0A182UZE0 A0A182TJD7 A0A182LEA2 A0A182XAI2 A0A182HSC2 Q7QCJ9 A0A182NR95 A0A1Q3F109 A0A182RQL5 A0A182Y6R0 Q171Y8 A0A023EMR0 A0A1B6HUY9 A0A1B6JYM1 A0A0C9Q9R9 A0A182FAG8 A0A2R5LG55 A0A293LNH3 B7PGI7 A0A0K8REF6 A0A226EJF6 Q9VNG0 B3P2I0 B4PUW2 A0A182SJA2 A0A2S2QND0 B4I453 B4JEV6 A0A3B0KPZ9 B4K8H3 A0A0L0BXT6 A0A0M4EP60 B4QX01 B3LWP3 B4M6L8 B5DXU2 B4G5L8 A0A1I8NEW0 A0A224YT23 A0A1W4UWI5 A0A131YN01 B4NG68 L7ME82 A0A1B0AWK8 A0A131XBG5 A0A2H8TX07 A0A2S2PNP7
A0A194PQB1 D2X8R5 A0A067R5Y6 A0A151IRJ9 A0A151IBA3 A0A195BTT5 A0A158NNR1 F4WV32 A0A195EW54 A0A151WSW2 A0A154PJQ4 A0A0M8ZVA6 A0A0L7R9J9 A0A026W257 A0A023F1U2 A0A224XUV3 A0A069DS88 E2C2M0 A0A2A3EJZ2 A0A087ZUD9 E0VTY9 A0A0C9RRE4 E2A3F7 A0A0A9WHB3 A0A0T6AYR4 A0A1Y1LMM7 A0A1B0CH03 A0A1B0DFY2 D6WH39 A0A1L8DFL3 K7J937 A0A182IS05 A0A232EZH6 A0A084W710 J3JUN3 A0A2M4AUD7 A0A182K701 W5JVP5 B0X296 A0A182P3V8 A0A182QAA0 A0A2J7QWU9 A0A182MLT5 A0A182VZY3 A0A2P8XYT8 A0A182UZE0 A0A182TJD7 A0A182LEA2 A0A182XAI2 A0A182HSC2 Q7QCJ9 A0A182NR95 A0A1Q3F109 A0A182RQL5 A0A182Y6R0 Q171Y8 A0A023EMR0 A0A1B6HUY9 A0A1B6JYM1 A0A0C9Q9R9 A0A182FAG8 A0A2R5LG55 A0A293LNH3 B7PGI7 A0A0K8REF6 A0A226EJF6 Q9VNG0 B3P2I0 B4PUW2 A0A182SJA2 A0A2S2QND0 B4I453 B4JEV6 A0A3B0KPZ9 B4K8H3 A0A0L0BXT6 A0A0M4EP60 B4QX01 B3LWP3 B4M6L8 B5DXU2 B4G5L8 A0A1I8NEW0 A0A224YT23 A0A1W4UWI5 A0A131YN01 B4NG68 L7ME82 A0A1B0AWK8 A0A131XBG5 A0A2H8TX07 A0A2S2PNP7
Pubmed
19121390
22118469
26227816
26354079
28756777
20062520
+ More
24845553 21347285 21719571 24508170 30249741 25474469 26334808 20798317 20566863 25401762 26823975 28004739 18362917 19820115 20075255 28648823 24438588 22516182 23537049 20920257 23761445 29403074 20966253 12364791 25244985 17510324 24945155 26483478 10731132 12537572 11259581 12021283 16751183 17994087 17550304 26108605 15632085 25315136 28797301 26830274 25576852 28049606
24845553 21347285 21719571 24508170 30249741 25474469 26334808 20798317 20566863 25401762 26823975 28004739 18362917 19820115 20075255 28648823 24438588 22516182 23537049 20920257 23761445 29403074 20966253 12364791 25244985 17510324 24945155 26483478 10731132 12537572 11259581 12021283 16751183 17994087 17550304 26108605 15632085 25315136 28797301 26830274 25576852 28049606
EMBL
BABH01027344
AGBW02013553
OWR43221.1
JTDY01000494
KOB76920.1
KQ460970
+ More
KPJ10202.1 KZ150156 PZC72783.1 ODYU01003405 SOQ42118.1 KQ459597 KPI95153.1 GU058050 ACY25396.1 KK852680 KDR18601.1 KQ981124 KYN09322.1 KQ978169 KYM96595.1 KQ976417 KYM89834.1 ADTU01021728 GL888384 EGI61920.1 KQ981953 KYN32500.1 KQ982766 KYQ50970.1 KQ434938 KZC12053.1 KQ435867 KOX70259.1 KQ414627 KOC67431.1 KK107485 QOIP01000006 EZA50098.1 RLU21457.1 GBBI01003220 JAC15492.1 GFTR01004667 JAW11759.1 GBGD01002367 JAC86522.1 GL452187 EFN77798.1 KZ288223 PBC32010.1 DS235777 EEB16845.1 GBYB01011119 GBYB01011120 JAG80886.1 JAG80887.1 GL436428 EFN72021.1 GBHO01035767 GBRD01013153 GDHC01013892 JAG07837.1 JAG52673.1 JAQ04737.1 LJIG01022494 KRT80281.1 GEZM01057549 JAV72257.1 AJWK01011791 AJVK01059119 KQ971321 EFA00114.1 GFDF01008846 JAV05238.1 AAZX01010347 AAZX01016424 NNAY01001474 OXU23842.1 ATLV01021084 KE525312 KFB46004.1 APGK01057723 BT126948 KB741280 KB632059 AEE61910.1 ENN71023.1 ERL88383.1 GGFK01011011 MBW44332.1 ADMH02000180 ETN67478.1 DS232283 EDS39119.1 AXCN02001467 NEVH01009418 PNF33061.1 AXCM01000211 PYGN01001149 PSN37167.1 APCN01000261 AAAB01008859 GFDL01013821 JAV21224.1 CH477443 JXUM01017361 GAPW01002955 KQ560481 JAC10643.1 KXJ82199.1 GECU01029207 JAS78499.1 GECU01003399 JAT04308.1 GBYB01011118 JAG80885.1 GGLE01004323 MBY08449.1 GFWV01005543 MAA30273.1 ABJB010048520 ABJB010250074 ABJB010376046 ABJB010497670 ABJB010518074 ABJB010544081 ABJB010682456 ABJB010783117 ABJB010801497 ABJB010861705 DS708364 EEC05709.1 GADI01004261 JAA69547.1 LNIX01000003 OXA57853.1 AE014297 AY075500 CH954181 EDV47930.1 CM000160 EDW97769.1 GGMS01009998 MBY79201.1 CH480821 EDW54996.1 CH916369 EDV93237.1 OUUW01000013 SPP87916.1 CH933806 EDW14372.2 JRES01001168 KNC24863.1 CP012526 ALC47527.1 CM000364 EDX11763.1 CH902617 EDV41637.1 CH940652 EDW59294.2 CM000070 EDY67908.1 CH479179 EDW24884.1 GFPF01009622 MAA20768.1 GEDV01008635 JAP79922.1 CH964251 EDW83285.1 GACK01002503 JAA62531.1 JXJN01004812 GEFH01003587 JAP64994.1 GFXV01006971 MBW18776.1 GGMR01017847 MBY30466.1
KPJ10202.1 KZ150156 PZC72783.1 ODYU01003405 SOQ42118.1 KQ459597 KPI95153.1 GU058050 ACY25396.1 KK852680 KDR18601.1 KQ981124 KYN09322.1 KQ978169 KYM96595.1 KQ976417 KYM89834.1 ADTU01021728 GL888384 EGI61920.1 KQ981953 KYN32500.1 KQ982766 KYQ50970.1 KQ434938 KZC12053.1 KQ435867 KOX70259.1 KQ414627 KOC67431.1 KK107485 QOIP01000006 EZA50098.1 RLU21457.1 GBBI01003220 JAC15492.1 GFTR01004667 JAW11759.1 GBGD01002367 JAC86522.1 GL452187 EFN77798.1 KZ288223 PBC32010.1 DS235777 EEB16845.1 GBYB01011119 GBYB01011120 JAG80886.1 JAG80887.1 GL436428 EFN72021.1 GBHO01035767 GBRD01013153 GDHC01013892 JAG07837.1 JAG52673.1 JAQ04737.1 LJIG01022494 KRT80281.1 GEZM01057549 JAV72257.1 AJWK01011791 AJVK01059119 KQ971321 EFA00114.1 GFDF01008846 JAV05238.1 AAZX01010347 AAZX01016424 NNAY01001474 OXU23842.1 ATLV01021084 KE525312 KFB46004.1 APGK01057723 BT126948 KB741280 KB632059 AEE61910.1 ENN71023.1 ERL88383.1 GGFK01011011 MBW44332.1 ADMH02000180 ETN67478.1 DS232283 EDS39119.1 AXCN02001467 NEVH01009418 PNF33061.1 AXCM01000211 PYGN01001149 PSN37167.1 APCN01000261 AAAB01008859 GFDL01013821 JAV21224.1 CH477443 JXUM01017361 GAPW01002955 KQ560481 JAC10643.1 KXJ82199.1 GECU01029207 JAS78499.1 GECU01003399 JAT04308.1 GBYB01011118 JAG80885.1 GGLE01004323 MBY08449.1 GFWV01005543 MAA30273.1 ABJB010048520 ABJB010250074 ABJB010376046 ABJB010497670 ABJB010518074 ABJB010544081 ABJB010682456 ABJB010783117 ABJB010801497 ABJB010861705 DS708364 EEC05709.1 GADI01004261 JAA69547.1 LNIX01000003 OXA57853.1 AE014297 AY075500 CH954181 EDV47930.1 CM000160 EDW97769.1 GGMS01009998 MBY79201.1 CH480821 EDW54996.1 CH916369 EDV93237.1 OUUW01000013 SPP87916.1 CH933806 EDW14372.2 JRES01001168 KNC24863.1 CP012526 ALC47527.1 CM000364 EDX11763.1 CH902617 EDV41637.1 CH940652 EDW59294.2 CM000070 EDY67908.1 CH479179 EDW24884.1 GFPF01009622 MAA20768.1 GEDV01008635 JAP79922.1 CH964251 EDW83285.1 GACK01002503 JAA62531.1 JXJN01004812 GEFH01003587 JAP64994.1 GFXV01006971 MBW18776.1 GGMR01017847 MBY30466.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000053240
UP000053268
UP000027135
+ More
UP000078492 UP000078542 UP000078540 UP000005205 UP000007755 UP000078541 UP000075809 UP000076502 UP000053105 UP000053825 UP000053097 UP000279307 UP000008237 UP000242457 UP000005203 UP000009046 UP000000311 UP000092461 UP000092462 UP000007266 UP000002358 UP000075880 UP000215335 UP000030765 UP000019118 UP000030742 UP000075881 UP000000673 UP000002320 UP000075885 UP000075886 UP000235965 UP000075883 UP000075920 UP000245037 UP000075903 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000075884 UP000075900 UP000076408 UP000008820 UP000069940 UP000249989 UP000069272 UP000001555 UP000198287 UP000000803 UP000008711 UP000002282 UP000075901 UP000001292 UP000001070 UP000268350 UP000009192 UP000037069 UP000092553 UP000000304 UP000007801 UP000008792 UP000001819 UP000008744 UP000095301 UP000192221 UP000007798 UP000092460
UP000078492 UP000078542 UP000078540 UP000005205 UP000007755 UP000078541 UP000075809 UP000076502 UP000053105 UP000053825 UP000053097 UP000279307 UP000008237 UP000242457 UP000005203 UP000009046 UP000000311 UP000092461 UP000092462 UP000007266 UP000002358 UP000075880 UP000215335 UP000030765 UP000019118 UP000030742 UP000075881 UP000000673 UP000002320 UP000075885 UP000075886 UP000235965 UP000075883 UP000075920 UP000245037 UP000075903 UP000075902 UP000075882 UP000076407 UP000075840 UP000007062 UP000075884 UP000075900 UP000076408 UP000008820 UP000069940 UP000249989 UP000069272 UP000001555 UP000198287 UP000000803 UP000008711 UP000002282 UP000075901 UP000001292 UP000001070 UP000268350 UP000009192 UP000037069 UP000092553 UP000000304 UP000007801 UP000008792 UP000001819 UP000008744 UP000095301 UP000192221 UP000007798 UP000092460
Interpro
ProteinModelPortal
H9J8C6
A0A212EP38
A0A0L7LNA8
A0A0N1INM8
A0A2W1BM73
A0A2H1VMT2
+ More
A0A194PQB1 D2X8R5 A0A067R5Y6 A0A151IRJ9 A0A151IBA3 A0A195BTT5 A0A158NNR1 F4WV32 A0A195EW54 A0A151WSW2 A0A154PJQ4 A0A0M8ZVA6 A0A0L7R9J9 A0A026W257 A0A023F1U2 A0A224XUV3 A0A069DS88 E2C2M0 A0A2A3EJZ2 A0A087ZUD9 E0VTY9 A0A0C9RRE4 E2A3F7 A0A0A9WHB3 A0A0T6AYR4 A0A1Y1LMM7 A0A1B0CH03 A0A1B0DFY2 D6WH39 A0A1L8DFL3 K7J937 A0A182IS05 A0A232EZH6 A0A084W710 J3JUN3 A0A2M4AUD7 A0A182K701 W5JVP5 B0X296 A0A182P3V8 A0A182QAA0 A0A2J7QWU9 A0A182MLT5 A0A182VZY3 A0A2P8XYT8 A0A182UZE0 A0A182TJD7 A0A182LEA2 A0A182XAI2 A0A182HSC2 Q7QCJ9 A0A182NR95 A0A1Q3F109 A0A182RQL5 A0A182Y6R0 Q171Y8 A0A023EMR0 A0A1B6HUY9 A0A1B6JYM1 A0A0C9Q9R9 A0A182FAG8 A0A2R5LG55 A0A293LNH3 B7PGI7 A0A0K8REF6 A0A226EJF6 Q9VNG0 B3P2I0 B4PUW2 A0A182SJA2 A0A2S2QND0 B4I453 B4JEV6 A0A3B0KPZ9 B4K8H3 A0A0L0BXT6 A0A0M4EP60 B4QX01 B3LWP3 B4M6L8 B5DXU2 B4G5L8 A0A1I8NEW0 A0A224YT23 A0A1W4UWI5 A0A131YN01 B4NG68 L7ME82 A0A1B0AWK8 A0A131XBG5 A0A2H8TX07 A0A2S2PNP7
A0A194PQB1 D2X8R5 A0A067R5Y6 A0A151IRJ9 A0A151IBA3 A0A195BTT5 A0A158NNR1 F4WV32 A0A195EW54 A0A151WSW2 A0A154PJQ4 A0A0M8ZVA6 A0A0L7R9J9 A0A026W257 A0A023F1U2 A0A224XUV3 A0A069DS88 E2C2M0 A0A2A3EJZ2 A0A087ZUD9 E0VTY9 A0A0C9RRE4 E2A3F7 A0A0A9WHB3 A0A0T6AYR4 A0A1Y1LMM7 A0A1B0CH03 A0A1B0DFY2 D6WH39 A0A1L8DFL3 K7J937 A0A182IS05 A0A232EZH6 A0A084W710 J3JUN3 A0A2M4AUD7 A0A182K701 W5JVP5 B0X296 A0A182P3V8 A0A182QAA0 A0A2J7QWU9 A0A182MLT5 A0A182VZY3 A0A2P8XYT8 A0A182UZE0 A0A182TJD7 A0A182LEA2 A0A182XAI2 A0A182HSC2 Q7QCJ9 A0A182NR95 A0A1Q3F109 A0A182RQL5 A0A182Y6R0 Q171Y8 A0A023EMR0 A0A1B6HUY9 A0A1B6JYM1 A0A0C9Q9R9 A0A182FAG8 A0A2R5LG55 A0A293LNH3 B7PGI7 A0A0K8REF6 A0A226EJF6 Q9VNG0 B3P2I0 B4PUW2 A0A182SJA2 A0A2S2QND0 B4I453 B4JEV6 A0A3B0KPZ9 B4K8H3 A0A0L0BXT6 A0A0M4EP60 B4QX01 B3LWP3 B4M6L8 B5DXU2 B4G5L8 A0A1I8NEW0 A0A224YT23 A0A1W4UWI5 A0A131YN01 B4NG68 L7ME82 A0A1B0AWK8 A0A131XBG5 A0A2H8TX07 A0A2S2PNP7
Ontologies
GO
PANTHER
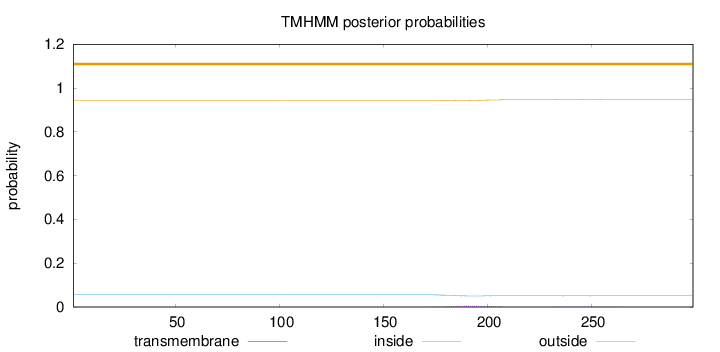
Topology
Subcellular location
Nucleus
Length:
299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15065
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05593
outside
1 - 299
Population Genetic Test Statistics
Pi
17.82328
Theta
17.638295
Tajima's D
0.031745
CLR
2.031893
CSRT
0.384030798460077
Interpretation
Uncertain
