Gene
KWMTBOMO03109
Pre Gene Modal
BGIBMGA005801
Annotation
nicotinic_acetylcholine_receptor_subunit_alpha_6_mRNA_transcript_variant_2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.807
Sequence
CDS
ATGATTGATGAAGGGAAACGGAACAAATTTCAAATTCAGACGATTGTTCTTGTTCTACCAGTGTCGGAGCAAGGTCCACACGAAAAACGTCTGCTGAACGCCCTGCTGGCGTCTTACAACACCCTGGAGCGTCCTGTGGCCAATGAGAGCGAGCCCCTAGAGGTACGCTTTGGGCTGACACTGCAACAAATCATAGATGTGGACGAGAAGAATCAACTACTTATAACCAATATATGGCTGTCATTGGAATGGAATGACTACAACCTAAGGTGGAACGAGAGCGAGTACGGAGGCGTCAAGGACTTAAGGATAACACCGAACAAACTTTGGAAACCAGACGTTCTCATGTACAACAGTGCTGACGAGGGTTTTGACGGGACATACCAGACGAACGTGGTGGTCAGAAACAACGGCAGTTGTCTGTACGTGCCCCCGGGCATTTTCAAGAGCACATGCAAGATAGATATCACGTGGTTTCCCTTCGACGACCAACACTGTGATATGAAGTTTGGTAGCTGGACATATGATGGCAATCAGCTGGACCTCATCTTGAAGGACGAAGCCGGTGGCGACCTATCAGATTTCATCACGAATGGAGAATGGTACTTAATAGGAATGCCGGGCAAAAAGAACACAATTACATACGCTTGCTGCCCAGAGCCGTACGTGGACGTCACCTTCACCATCATGATCAGGAGGAGAACGCTGTACTACTTCTTCAATCTCATCGTGCCGTGCGTTCTGATATCCTCAATGGCGCTACTAGGCTTCACGCTACCACCAGATTCTGGGGAGAAATTGACACTCGGAGTCACTATTCTACTGTCGCTGACGGTGTTTCTCAATCTGGTGGCAGAAACTCTGCCGCAGGTGTCCGATGCTATCCCCTTGTTAGGTACATATTTCAACTGTATTATGTTCATGGTGGCCTCGTCTGTGGTGCTCACCGTTGTGGTGTTGAACTACCATCATCGAACGGCTGACATCCATGAGATGCCACAATGGATAAAGACTGTGTTCCTCCAATGGCTGCCCTGGATTCTGCGTATGTCCAGGCCGGGTAAGAAGATCACGAGGAAGACCATCATGATGTCCAACAGGATGCGAGAGTTGGAGCTAAAAGAGCGTTCGTCTAAGTCGCTGTTAGCCAACGTGCTGGATATAGACGACGACTTTAGACATGCCCCGCCCCCGCCCAACAGTACTGCTTCTACAGGCAACTTGGGGCCTGGAACGGACTTTCGGCGTTCGTTCGTCCGCCCTTCGACGATGGAGGACGTGGGCGGAGGGCTCAGTAGCCACCACAGGGAGCTGCATCTCATCCTGCGCGAACTCCAGTTCATCACAGCCAGGATGAAGAAGGCCGATGAGGAGGCCGAGCTGATCAGCGACTGGAAATTCGCTGCAATGGTAGTGGACAGGTTTTGCTTGTTCGTGTTCACGTTGTTCACGATAATAGCGACGGTGGCCGTGCTGTTGTCGGCGCCGCATATCATCGTGCAGTGA
Protein
MIDEGKRNKFQIQTIVLVLPVSEQGPHEKRLLNALLASYNTLERPVANESEPLEVRFGLTLQQIIDVDEKNQLLITNIWLSLEWNDYNLRWNESEYGGVKDLRITPNKLWKPDVLMYNSADEGFDGTYQTNVVVRNNGSCLYVPPGIFKSTCKIDITWFPFDDQHCDMKFGSWTYDGNQLDLILKDEAGGDLSDFITNGEWYLIGMPGKKNTITYACCPEPYVDVTFTIMIRRRTLYYFFNLIVPCVLISSMALLGFTLPPDSGEKLTLGVTILLSLTVFLNLVAETLPQVSDAIPLLGTYFNCIMFMVASSVVLTVVVLNYHHRTADIHEMPQWIKTVFLQWLPWILRMSRPGKKITRKTIMMSNRMRELELKERSSKSLLANVLDIDDDFRHAPPPPNSTASTGNLGPGTDFRRSFVRPSTMEDVGGGLSSHHRELHLILRELQFITARMKKADEEAELISDWKFAAMVVDRFCLFVFTLFTIIATVAVLLSAPHIIVQ
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
A8CGL6
A1EGV5
A5A2M6
A0A2A4JGH6
A0A2W1BAC7
A1EGV4
+ More
A0A0H4U7U0 Q9XZI4 A1EGV6 D8UUW8 A0A0H4TMF2 A0A0S1TL21 A0A0A7RQA0 A0A346IHV1 D8UWB3 D3JL32 A8CGM0 A0A140CTI3 D9YT57 D9YT60 D2X8R6 D9YT59 D9YT61 D9YT62 A0A139WNV9 C3TS36 A0A1S4FID0 A8DIT3 A0A0P8XXV7 C3TS34 A0A0R3NRK7 A0A1W4W8N4 A0A0J9R0C5 Q8IPE2 A0A0R1DNB2 A0A0Q5WLY3 A0A0Q9X712 C3TS35 A0A0Q9W8H3 Q7PTL9 M4H3L1 A0A0P9BRB7 Q8T7S1 A0A0R3NX37 A0A1W4VXP1 C3TS31 C3TS37 G9CU48 A8DIS9 A0A0J9QZ20 A0A0Q9XHQ5 A0A0R1DM16 A0A0P8ZHV2 B4N125 A0A0Q5WME6 Q9VL79 A0A222C753 Q16ZU4 A0A0R3NRJ6 A0A0Q9W8J0 A0A0Q9X676 A0A0J9QZJ7 A0A0N8NZG8 A0A0Q5WAA2 Q7KTF9 A0A0R1DL23 W8BEN9 A5H033 M4H3G0 G9E3J3 A0A0K0NTY9 A8DIT5 A0A1W4VXP6 A0A0R3NS12 A5H032 A0A0J9TJR6 Q7KTF7 B4NYM5 A0A0Q5WJ74 M4H3Y7 A0A1I8NYF7 A0A0P8XHK1 K4JH01 A0A1W4VXN1 A5H031 Q8T7S2 B4LQB0 A0A0R3NRG3 Q86MN8 A0A0P8Y5B5 A0A0Q9XF72 A0A0R3NWT4 Q6NNY5 A0A0J9QZI0 M9PFD8 A0A0R1DKV4 A0A0Q5WJ24 Q8T7S0 A0A0Q9W8K3 K4JF52
A0A0H4U7U0 Q9XZI4 A1EGV6 D8UUW8 A0A0H4TMF2 A0A0S1TL21 A0A0A7RQA0 A0A346IHV1 D8UWB3 D3JL32 A8CGM0 A0A140CTI3 D9YT57 D9YT60 D2X8R6 D9YT59 D9YT61 D9YT62 A0A139WNV9 C3TS36 A0A1S4FID0 A8DIT3 A0A0P8XXV7 C3TS34 A0A0R3NRK7 A0A1W4W8N4 A0A0J9R0C5 Q8IPE2 A0A0R1DNB2 A0A0Q5WLY3 A0A0Q9X712 C3TS35 A0A0Q9W8H3 Q7PTL9 M4H3L1 A0A0P9BRB7 Q8T7S1 A0A0R3NX37 A0A1W4VXP1 C3TS31 C3TS37 G9CU48 A8DIS9 A0A0J9QZ20 A0A0Q9XHQ5 A0A0R1DM16 A0A0P8ZHV2 B4N125 A0A0Q5WME6 Q9VL79 A0A222C753 Q16ZU4 A0A0R3NRJ6 A0A0Q9W8J0 A0A0Q9X676 A0A0J9QZJ7 A0A0N8NZG8 A0A0Q5WAA2 Q7KTF9 A0A0R1DL23 W8BEN9 A5H033 M4H3G0 G9E3J3 A0A0K0NTY9 A8DIT5 A0A1W4VXP6 A0A0R3NS12 A5H032 A0A0J9TJR6 Q7KTF7 B4NYM5 A0A0Q5WJ74 M4H3Y7 A0A1I8NYF7 A0A0P8XHK1 K4JH01 A0A1W4VXN1 A5H031 Q8T7S2 B4LQB0 A0A0R3NRG3 Q86MN8 A0A0P8Y5B5 A0A0Q9XF72 A0A0R3NWT4 Q6NNY5 A0A0J9QZI0 M9PFD8 A0A0R1DKV4 A0A0Q5WJ24 Q8T7S0 A0A0Q9W8K3 K4JF52
Pubmed
17868469
17597521
28756777
25504620
20062520
26855198
+ More
18362917 19820115 19320762 17510324 17880682 17994087 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12364791 14747013 15676276 17210077 22898623 11973307 26109357 26109356 24495485 17439546
18362917 19820115 19320762 17510324 17880682 17994087 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12364791 14747013 15676276 17210077 22898623 11973307 26109357 26109356 24495485 17439546
EMBL
EU082082
ABV45518.1
EF127798
ABL67935.1
EF521820
ABP96888.1
+ More
NWSH01001615 PCG70684.1 KZ150420 PZC70934.1 EF127797 ABL67934.1 KP711048 AKQ12744.1 AF143847 AAD32698.1 EF127799 ABL67936.1 GQ228552 ADD69773.1 KP711055 AKQ12751.1 KP771859 ALM23508.1 KP101245 AJA39821.1 MH138165 AXP17106.1 GQ247883 ADE34345.1 GU207835 KU130399 ADB84598.1 AMH87606.1 EU082083 ABV45519.1 KU130400 AMH87607.1 GU060293 ADC29632.1 GU060296 ADC29635.1 GU058050 ACY25395.1 GU060295 ADC29634.1 GU060297 ADC29636.1 GU060298 ADC29637.1 KQ971307 KYB29612.1 EU930066 ACM09861.1 EF526087 ABS86909.1 KYB29613.1 CH902624 KPU74286.1 EU930064 ACM09859.1 CH379060 KRT03800.1 CM002910 KMY89339.1 AE014134 AAN10709.2 CM000157 KRJ97981.1 CH954177 KQS70399.1 CH933807 KRG03749.1 EU930065 ACM09860.1 CH940649 KRF81069.1 AY705400 AAAB01008799 AAU12509.1 EAA03696.4 JN560169 AFN88982.1 KPU74283.1 AF321447 AAM13394.1 KRT03805.1 EU930061 ACM09856.1 EU930067 ACM09862.1 JF731243 KP725465 AEA40429.1 AKV94622.1 EF526086 EU930068 ABS86908.1 ACM09863.1 KYB29614.1 KMY89337.1 KRG03751.1 KRJ97982.1 KPU74285.1 CH963920 EDW78187.2 KQS70394.1 AF321445 AAF52817.3 AAM13392.1 KX525264 ASO96849.1 CH477484 EAT40177.1 KRT03806.1 KRF81066.1 KRG03750.1 KMY89343.1 KPU74284.1 KQS70397.1 AAS64670.1 ADV37014.1 KRJ97980.1 KRJ97983.1 GAMC01006785 JAB99770.1 DQ498131 ABJ09670.1 JN560168 AFN88981.1 JF974072 AEN03030.1 KP260561 AKO69818.1 EF526088 ABS86910.1 KYB29611.1 KRT03801.1 DQ498130 ABJ09669.1 KMY89335.1 AAS64672.1 EDW88689.2 KQS70398.1 JN560167 AFN88980.1 KPU74281.1 JX466887 AFU82558.1 DQ498129 ABJ09668.1 AF321446 AAM13393.1 EDW63360.2 KRT03809.1 AJ554209 CAD86935.1 KPU74282.1 KRG03748.1 KRT03807.1 BT011147 AAR82815.1 KMY89341.1 AGB92837.1 KRJ97979.1 KQS70395.1 AF321448 AAM13395.1 KRF81068.1 JX466888 AFU82559.1
NWSH01001615 PCG70684.1 KZ150420 PZC70934.1 EF127797 ABL67934.1 KP711048 AKQ12744.1 AF143847 AAD32698.1 EF127799 ABL67936.1 GQ228552 ADD69773.1 KP711055 AKQ12751.1 KP771859 ALM23508.1 KP101245 AJA39821.1 MH138165 AXP17106.1 GQ247883 ADE34345.1 GU207835 KU130399 ADB84598.1 AMH87606.1 EU082083 ABV45519.1 KU130400 AMH87607.1 GU060293 ADC29632.1 GU060296 ADC29635.1 GU058050 ACY25395.1 GU060295 ADC29634.1 GU060297 ADC29636.1 GU060298 ADC29637.1 KQ971307 KYB29612.1 EU930066 ACM09861.1 EF526087 ABS86909.1 KYB29613.1 CH902624 KPU74286.1 EU930064 ACM09859.1 CH379060 KRT03800.1 CM002910 KMY89339.1 AE014134 AAN10709.2 CM000157 KRJ97981.1 CH954177 KQS70399.1 CH933807 KRG03749.1 EU930065 ACM09860.1 CH940649 KRF81069.1 AY705400 AAAB01008799 AAU12509.1 EAA03696.4 JN560169 AFN88982.1 KPU74283.1 AF321447 AAM13394.1 KRT03805.1 EU930061 ACM09856.1 EU930067 ACM09862.1 JF731243 KP725465 AEA40429.1 AKV94622.1 EF526086 EU930068 ABS86908.1 ACM09863.1 KYB29614.1 KMY89337.1 KRG03751.1 KRJ97982.1 KPU74285.1 CH963920 EDW78187.2 KQS70394.1 AF321445 AAF52817.3 AAM13392.1 KX525264 ASO96849.1 CH477484 EAT40177.1 KRT03806.1 KRF81066.1 KRG03750.1 KMY89343.1 KPU74284.1 KQS70397.1 AAS64670.1 ADV37014.1 KRJ97980.1 KRJ97983.1 GAMC01006785 JAB99770.1 DQ498131 ABJ09670.1 JN560168 AFN88981.1 JF974072 AEN03030.1 KP260561 AKO69818.1 EF526088 ABS86910.1 KYB29611.1 KRT03801.1 DQ498130 ABJ09669.1 KMY89335.1 AAS64672.1 EDW88689.2 KQS70398.1 JN560167 AFN88980.1 KPU74281.1 JX466887 AFU82558.1 DQ498129 ABJ09668.1 AF321446 AAM13393.1 EDW63360.2 KRT03809.1 AJ554209 CAD86935.1 KPU74282.1 KRG03748.1 KRT03807.1 BT011147 AAR82815.1 KMY89341.1 AGB92837.1 KRJ97979.1 KQS70395.1 AF321448 AAM13395.1 KRF81068.1 JX466888 AFU82559.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A8CGL6
A1EGV5
A5A2M6
A0A2A4JGH6
A0A2W1BAC7
A1EGV4
+ More
A0A0H4U7U0 Q9XZI4 A1EGV6 D8UUW8 A0A0H4TMF2 A0A0S1TL21 A0A0A7RQA0 A0A346IHV1 D8UWB3 D3JL32 A8CGM0 A0A140CTI3 D9YT57 D9YT60 D2X8R6 D9YT59 D9YT61 D9YT62 A0A139WNV9 C3TS36 A0A1S4FID0 A8DIT3 A0A0P8XXV7 C3TS34 A0A0R3NRK7 A0A1W4W8N4 A0A0J9R0C5 Q8IPE2 A0A0R1DNB2 A0A0Q5WLY3 A0A0Q9X712 C3TS35 A0A0Q9W8H3 Q7PTL9 M4H3L1 A0A0P9BRB7 Q8T7S1 A0A0R3NX37 A0A1W4VXP1 C3TS31 C3TS37 G9CU48 A8DIS9 A0A0J9QZ20 A0A0Q9XHQ5 A0A0R1DM16 A0A0P8ZHV2 B4N125 A0A0Q5WME6 Q9VL79 A0A222C753 Q16ZU4 A0A0R3NRJ6 A0A0Q9W8J0 A0A0Q9X676 A0A0J9QZJ7 A0A0N8NZG8 A0A0Q5WAA2 Q7KTF9 A0A0R1DL23 W8BEN9 A5H033 M4H3G0 G9E3J3 A0A0K0NTY9 A8DIT5 A0A1W4VXP6 A0A0R3NS12 A5H032 A0A0J9TJR6 Q7KTF7 B4NYM5 A0A0Q5WJ74 M4H3Y7 A0A1I8NYF7 A0A0P8XHK1 K4JH01 A0A1W4VXN1 A5H031 Q8T7S2 B4LQB0 A0A0R3NRG3 Q86MN8 A0A0P8Y5B5 A0A0Q9XF72 A0A0R3NWT4 Q6NNY5 A0A0J9QZI0 M9PFD8 A0A0R1DKV4 A0A0Q5WJ24 Q8T7S0 A0A0Q9W8K3 K4JF52
A0A0H4U7U0 Q9XZI4 A1EGV6 D8UUW8 A0A0H4TMF2 A0A0S1TL21 A0A0A7RQA0 A0A346IHV1 D8UWB3 D3JL32 A8CGM0 A0A140CTI3 D9YT57 D9YT60 D2X8R6 D9YT59 D9YT61 D9YT62 A0A139WNV9 C3TS36 A0A1S4FID0 A8DIT3 A0A0P8XXV7 C3TS34 A0A0R3NRK7 A0A1W4W8N4 A0A0J9R0C5 Q8IPE2 A0A0R1DNB2 A0A0Q5WLY3 A0A0Q9X712 C3TS35 A0A0Q9W8H3 Q7PTL9 M4H3L1 A0A0P9BRB7 Q8T7S1 A0A0R3NX37 A0A1W4VXP1 C3TS31 C3TS37 G9CU48 A8DIS9 A0A0J9QZ20 A0A0Q9XHQ5 A0A0R1DM16 A0A0P8ZHV2 B4N125 A0A0Q5WME6 Q9VL79 A0A222C753 Q16ZU4 A0A0R3NRJ6 A0A0Q9W8J0 A0A0Q9X676 A0A0J9QZJ7 A0A0N8NZG8 A0A0Q5WAA2 Q7KTF9 A0A0R1DL23 W8BEN9 A5H033 M4H3G0 G9E3J3 A0A0K0NTY9 A8DIT5 A0A1W4VXP6 A0A0R3NS12 A5H032 A0A0J9TJR6 Q7KTF7 B4NYM5 A0A0Q5WJ74 M4H3Y7 A0A1I8NYF7 A0A0P8XHK1 K4JH01 A0A1W4VXN1 A5H031 Q8T7S2 B4LQB0 A0A0R3NRG3 Q86MN8 A0A0P8Y5B5 A0A0Q9XF72 A0A0R3NWT4 Q6NNY5 A0A0J9QZI0 M9PFD8 A0A0R1DKV4 A0A0Q5WJ24 Q8T7S0 A0A0Q9W8K3 K4JF52
PDB
4BOT
E-value=1.00787e-82,
Score=782
Ontologies
GO
PANTHER
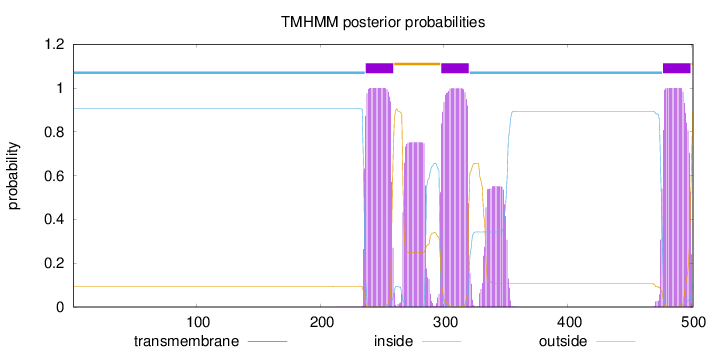
Topology
Subcellular location
Cell junction
Synapse
Postsynaptic cell membrane
Cell membrane
Synapse
Postsynaptic cell membrane
Cell membrane
Length:
501
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
92.6784600000001
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.90594
inside
1 - 236
TMhelix
237 - 259
outside
260 - 297
TMhelix
298 - 320
inside
321 - 476
TMhelix
477 - 499
outside
500 - 501
Population Genetic Test Statistics
Pi
29.672132
Theta
26.302772
Tajima's D
0.390458
CLR
1.125717
CSRT
0.486375681215939
Interpretation
Uncertain
