Gene
KWMTBOMO03097
Pre Gene Modal
BGIBMGA005763
Annotation
PREDICTED:_17-beta-hydroxysteroid_dehydrogenase_13-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.791 PlasmaMembrane Reliability : 1.807
Sequence
CDS
ATGGCGTCGGAACTAAGAACAGAAAAGGGGTCTTCTAATGGATTGACGGATAAAGCTCCCTGGACTGATGAGCAAGGCATAGTGATGAAGGTGTACCAAGCAGTGATGCTGACTCTGGAGATCGTGACCCTCTATATCAAGATGGTTCTCACCTGGTTCCATGCTATGTACCTGCTGGTAGTGCCCCCCGAGCCGAAGAGTGTCAATGGAGAGATTATCCTTATTACCGGCACAGGCCACGGCATGGGTAGGGAGACAGCGATGAGGTTTGCCAAGCTCGGAGGCGTGATAGTCTGTGTGGATATCAACCAGAAAACCAATCAGGAGACCGTGGACCTCATCAAGGAGCAGAAAGGAAAAGCGTATCGGTTCGAATGTGACGTCACAGACAGCGAGGCAGTGAAGGCTCTGGCCGAGAAGATCCGTCGTGAAGTAGGCGACGTTACCATGCTGGTGAACAACGCCGGCATCATGCCGTGCAAGCCGCTGCTGCAGCATGGGGAGGGGGAGATCAGGGCCGCCTTCAATGTCAATGTCATGGCTCATCTGTGGTTGATCCAAGCATTCCTACCCTCTATGATGGAGCGAAACCACGGTCACATCGTGGCGATGTCTTCGATGGCCGGTGTCCTTGGACTACGGAATTTAGTTCCATATTGCGCCACCAAGTTCGCTGTCAGAGGGCTAATGGAAGCGCTGCACGAAGAGTTAAGGGAGGACACGAGAGATTTCAGTGGCATCAAATTCACGGTGATCTGCCCGTACATAGTGGACACGGGACTGTGCAAGAACCCCAAGATCAAGTTCCCGTCGATGATGAAGATCCTGACGCCTGAAGAAGCTTCCGACTGCATCGTGGACGCGGTCCGGAGGAACTACTACGAGATCAGCATACCGACGTCGCTCTTATACGTCAACACTTTTTGCCGTCTATTGCCGAGGTCTGTGTCCTCGCATTTGAAGGATTTTTTGGATTCAGGACTTGAGGCACAGTAA
Protein
MASELRTEKGSSNGLTDKAPWTDEQGIVMKVYQAVMLTLEIVTLYIKMVLTWFHAMYLLVVPPEPKSVNGEIILITGTGHGMGRETAMRFAKLGGVIVCVDINQKTNQETVDLIKEQKGKAYRFECDVTDSEAVKALAEKIRREVGDVTMLVNNAGIMPCKPLLQHGEGEIRAAFNVNVMAHLWLIQAFLPSMMERNHGHIVAMSSMAGVLGLRNLVPYCATKFAVRGLMEALHEELREDTRDFSGIKFTVICPYIVDTGLCKNPKIKFPSMMKILTPEEASDCIVDAVRRNYYEISIPTSLLYVNTFCRLLPRSVSSHLKDFLDSGLEAQ
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J8C1
S4PKB0
A0A0N1PHH5
A0A212EHJ0
A0A2H1WWL5
H9J8G6
+ More
A0A212EHH8 A0A2A4K3Z4 A0A084VNW2 A0A0N1IP16 A0A2C9GPE1 Q7PRW2 A0A2H1VJQ9 A0A1Q3G5K6 A0A1B6C109 W5JLV1 B0WZ23 A0A1Q3FQL1 A0A182PSU8 A0A182RHK9 A0A182L878 A0A182QFY3 A0A182MHI6 A0A182TE30 A0A182YH69 A0A067QTW2 A0A182N8G2 A0A182W5M3 A0A182XBY7 A0A182UQG1 A0A1L8DKW9 A0A182IZ06 A0A1B6H6B3 A0A2J7PVM3 A0A1B6MJ99 A0A195D3V5 A0A2A4K4B3 A0A1B6GXZ2 A0A182FIS9 K7IU30 A0A224XGH6 Q16ZY5 A0A023EPJ1 E2C4N5 A0A0V0G980 F4W5L6 A0A0N7Z933 A0A158NSD4 A0A0L7QV45 A0A026WJP8 T1IBK9 A0A1B6D4X1 A0A161MZK5 A0A182GVC9 A0A023F371 A0A0P6J5T2 A0A023EPC0 B4NEJ5 A0A023EQF5 A0A146MDQ4 A0A182MN21 A0A146KSE4 A0A0K8T673 W8CBT6 F5HJM5 A0A2C9GPL9 A0A0A9XL74 Q9VKB8 A0A1W4VAZ0 B4P1Y1 A0A023ERM3 B3N3X7 A0A0J9R152 A0A336LVA3 B0WZ24 A0A0C9PL38 A0A034WN98 Q5BID6 A0A1Q3FII8 A0A182TBP2 E0W476 A0A0A1WXB0 A0A034WKG7 B4L823 A0A0K8VRM3 A0A2M4CXL7 A0A1I8PPZ8 D3TRJ0 B4N7A2 B3MVI2 A0A2M4BUP4 A0A2M4CXM9 A0A0K8UQP6 A0A2M3Z8X6 A0A0K8UNJ4 A0A1B0EX60 A0A1B0BV62 A0A1A9XFP3
A0A212EHH8 A0A2A4K3Z4 A0A084VNW2 A0A0N1IP16 A0A2C9GPE1 Q7PRW2 A0A2H1VJQ9 A0A1Q3G5K6 A0A1B6C109 W5JLV1 B0WZ23 A0A1Q3FQL1 A0A182PSU8 A0A182RHK9 A0A182L878 A0A182QFY3 A0A182MHI6 A0A182TE30 A0A182YH69 A0A067QTW2 A0A182N8G2 A0A182W5M3 A0A182XBY7 A0A182UQG1 A0A1L8DKW9 A0A182IZ06 A0A1B6H6B3 A0A2J7PVM3 A0A1B6MJ99 A0A195D3V5 A0A2A4K4B3 A0A1B6GXZ2 A0A182FIS9 K7IU30 A0A224XGH6 Q16ZY5 A0A023EPJ1 E2C4N5 A0A0V0G980 F4W5L6 A0A0N7Z933 A0A158NSD4 A0A0L7QV45 A0A026WJP8 T1IBK9 A0A1B6D4X1 A0A161MZK5 A0A182GVC9 A0A023F371 A0A0P6J5T2 A0A023EPC0 B4NEJ5 A0A023EQF5 A0A146MDQ4 A0A182MN21 A0A146KSE4 A0A0K8T673 W8CBT6 F5HJM5 A0A2C9GPL9 A0A0A9XL74 Q9VKB8 A0A1W4VAZ0 B4P1Y1 A0A023ERM3 B3N3X7 A0A0J9R152 A0A336LVA3 B0WZ24 A0A0C9PL38 A0A034WN98 Q5BID6 A0A1Q3FII8 A0A182TBP2 E0W476 A0A0A1WXB0 A0A034WKG7 B4L823 A0A0K8VRM3 A0A2M4CXL7 A0A1I8PPZ8 D3TRJ0 B4N7A2 B3MVI2 A0A2M4BUP4 A0A2M4CXM9 A0A0K8UQP6 A0A2M3Z8X6 A0A0K8UNJ4 A0A1B0EX60 A0A1B0BV62 A0A1A9XFP3
Pubmed
19121390
23622113
26354079
22118469
24438588
12364791
+ More
14747013 17210077 20920257 23761445 20966253 25244985 24845553 20075255 17510324 24945155 20798317 21719571 27129103 21347285 24508170 30249741 26483478 25474469 26999592 17994087 26823975 24495485 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 25348373 20566863 25830018 20353571 18057021
14747013 17210077 20920257 23761445 20966253 25244985 24845553 20075255 17510324 24945155 20798317 21719571 27129103 21347285 24508170 30249741 26483478 25474469 26999592 17994087 26823975 24495485 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 25348373 20566863 25830018 20353571 18057021
EMBL
BABH01027372
BABH01027373
BABH01027374
BABH01027375
BABH01027376
GAIX01004600
+ More
JAA87960.1 KQ460847 KPJ11975.1 AGBW02014879 OWR40948.1 ODYU01011214 SOQ56814.1 BABH01027377 AGBW02014880 OWR40937.1 NWSH01000175 PCG78746.1 ATLV01014931 KE524996 KFB39656.1 KPJ11976.1 APCN01001259 AAAB01008846 EAA06477.6 ODYU01002942 SOQ41079.1 GEYN01000189 JAV44800.1 GEDC01030244 GEDC01008499 JAS07054.1 JAS28799.1 ADMH02001200 ETN63754.1 DS232203 EDS37337.1 GFDL01005134 JAV29911.1 AXCN02002226 AXCM01002712 KK853021 KDR12387.1 GFDF01007094 JAV06990.1 GECU01037505 JAS70201.1 NEVH01020948 PNF20383.1 GEBQ01029339 GEBQ01024795 GEBQ01017340 GEBQ01003983 JAT10638.1 JAT15182.1 JAT22637.1 JAT35994.1 KQ976881 KYN07572.1 PCG78748.1 GECZ01002473 JAS67296.1 AAZX01011807 GFTR01004841 JAW11585.1 CH477480 EAT40239.1 GAPW01002186 JAC11412.1 GL452536 EFN77115.1 GECL01001744 JAP04380.1 GL887655 EGI70513.1 GDKW01001667 JAI54928.1 ADTU01024832 KQ414727 KOC62508.1 KK107167 QOIP01000010 EZA56272.1 RLU17457.1 ACPB03013753 GEDC01016578 GEDC01000925 JAS20720.1 JAS36373.1 GEMB01003343 JAR99871.1 JXUM01002260 JXUM01002261 KQ560135 KXJ84250.1 GBBI01003106 JAC15606.1 GDUN01000390 JAN95529.1 JXUM01020073 GAPW01002759 GAPW01002758 KQ560563 JAC10840.1 KXJ81885.1 CH964239 EDW82164.2 GAPW01002794 JAC10804.1 GDHC01001477 JAQ17152.1 GDHC01020487 JAP98141.1 GBRD01004802 GBRD01004801 JAG61019.1 GAMC01004536 JAC02020.1 EGK96486.1 GBHO01023203 JAG20401.1 AE014134 BT133278 AAF53158.3 AFC38909.1 AFH03671.1 AHN54381.1 CM000157 EDW88152.2 KRJ97607.1 GAPW01002264 JAC11334.1 CH954177 EDV58829.2 CM002910 KMY89871.1 UFQT01000115 SSX20267.1 EDS37338.1 GBYB01001718 JAG71485.1 GAKP01003317 JAC55635.1 BT021288 AAX33436.1 GFDL01007752 JAV27293.1 DS235886 EEB20432.1 GBXI01011234 GBXI01011183 JAD03058.1 JAD03109.1 GAKP01004709 JAC54243.1 CH933814 EDW05598.2 GDHF01010787 JAI41527.1 GGFL01005898 MBW70076.1 EZ424042 ADD20318.1 CH964182 EDW80243.2 CH902624 EDV33247.2 KPU74364.1 GGFJ01007646 MBW56787.1 GGFL01005899 MBW70077.1 GDHF01028530 GDHF01023295 GDHF01003485 JAI23784.1 JAI29019.1 JAI48829.1 GGFM01004127 MBW24878.1 GDHF01024226 GDHF01012080 JAI28088.1 JAI40234.1 AJVK01000821 JXJN01021143
JAA87960.1 KQ460847 KPJ11975.1 AGBW02014879 OWR40948.1 ODYU01011214 SOQ56814.1 BABH01027377 AGBW02014880 OWR40937.1 NWSH01000175 PCG78746.1 ATLV01014931 KE524996 KFB39656.1 KPJ11976.1 APCN01001259 AAAB01008846 EAA06477.6 ODYU01002942 SOQ41079.1 GEYN01000189 JAV44800.1 GEDC01030244 GEDC01008499 JAS07054.1 JAS28799.1 ADMH02001200 ETN63754.1 DS232203 EDS37337.1 GFDL01005134 JAV29911.1 AXCN02002226 AXCM01002712 KK853021 KDR12387.1 GFDF01007094 JAV06990.1 GECU01037505 JAS70201.1 NEVH01020948 PNF20383.1 GEBQ01029339 GEBQ01024795 GEBQ01017340 GEBQ01003983 JAT10638.1 JAT15182.1 JAT22637.1 JAT35994.1 KQ976881 KYN07572.1 PCG78748.1 GECZ01002473 JAS67296.1 AAZX01011807 GFTR01004841 JAW11585.1 CH477480 EAT40239.1 GAPW01002186 JAC11412.1 GL452536 EFN77115.1 GECL01001744 JAP04380.1 GL887655 EGI70513.1 GDKW01001667 JAI54928.1 ADTU01024832 KQ414727 KOC62508.1 KK107167 QOIP01000010 EZA56272.1 RLU17457.1 ACPB03013753 GEDC01016578 GEDC01000925 JAS20720.1 JAS36373.1 GEMB01003343 JAR99871.1 JXUM01002260 JXUM01002261 KQ560135 KXJ84250.1 GBBI01003106 JAC15606.1 GDUN01000390 JAN95529.1 JXUM01020073 GAPW01002759 GAPW01002758 KQ560563 JAC10840.1 KXJ81885.1 CH964239 EDW82164.2 GAPW01002794 JAC10804.1 GDHC01001477 JAQ17152.1 GDHC01020487 JAP98141.1 GBRD01004802 GBRD01004801 JAG61019.1 GAMC01004536 JAC02020.1 EGK96486.1 GBHO01023203 JAG20401.1 AE014134 BT133278 AAF53158.3 AFC38909.1 AFH03671.1 AHN54381.1 CM000157 EDW88152.2 KRJ97607.1 GAPW01002264 JAC11334.1 CH954177 EDV58829.2 CM002910 KMY89871.1 UFQT01000115 SSX20267.1 EDS37338.1 GBYB01001718 JAG71485.1 GAKP01003317 JAC55635.1 BT021288 AAX33436.1 GFDL01007752 JAV27293.1 DS235886 EEB20432.1 GBXI01011234 GBXI01011183 JAD03058.1 JAD03109.1 GAKP01004709 JAC54243.1 CH933814 EDW05598.2 GDHF01010787 JAI41527.1 GGFL01005898 MBW70076.1 EZ424042 ADD20318.1 CH964182 EDW80243.2 CH902624 EDV33247.2 KPU74364.1 GGFJ01007646 MBW56787.1 GGFL01005899 MBW70077.1 GDHF01028530 GDHF01023295 GDHF01003485 JAI23784.1 JAI29019.1 JAI48829.1 GGFM01004127 MBW24878.1 GDHF01024226 GDHF01012080 JAI28088.1 JAI40234.1 AJVK01000821 JXJN01021143
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000030765
UP000075840
+ More
UP000007062 UP000000673 UP000002320 UP000075885 UP000075900 UP000075882 UP000075886 UP000075883 UP000075902 UP000076408 UP000027135 UP000075884 UP000075920 UP000076407 UP000075903 UP000075880 UP000235965 UP000078542 UP000069272 UP000002358 UP000008820 UP000008237 UP000007755 UP000005205 UP000053825 UP000053097 UP000279307 UP000015103 UP000069940 UP000249989 UP000007798 UP000000803 UP000192221 UP000002282 UP000008711 UP000075901 UP000009046 UP000009192 UP000095300 UP000007801 UP000092462 UP000092460 UP000092443
UP000007062 UP000000673 UP000002320 UP000075885 UP000075900 UP000075882 UP000075886 UP000075883 UP000075902 UP000076408 UP000027135 UP000075884 UP000075920 UP000076407 UP000075903 UP000075880 UP000235965 UP000078542 UP000069272 UP000002358 UP000008820 UP000008237 UP000007755 UP000005205 UP000053825 UP000053097 UP000279307 UP000015103 UP000069940 UP000249989 UP000007798 UP000000803 UP000192221 UP000002282 UP000008711 UP000075901 UP000009046 UP000009192 UP000095300 UP000007801 UP000092462 UP000092460 UP000092443
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J8C1
S4PKB0
A0A0N1PHH5
A0A212EHJ0
A0A2H1WWL5
H9J8G6
+ More
A0A212EHH8 A0A2A4K3Z4 A0A084VNW2 A0A0N1IP16 A0A2C9GPE1 Q7PRW2 A0A2H1VJQ9 A0A1Q3G5K6 A0A1B6C109 W5JLV1 B0WZ23 A0A1Q3FQL1 A0A182PSU8 A0A182RHK9 A0A182L878 A0A182QFY3 A0A182MHI6 A0A182TE30 A0A182YH69 A0A067QTW2 A0A182N8G2 A0A182W5M3 A0A182XBY7 A0A182UQG1 A0A1L8DKW9 A0A182IZ06 A0A1B6H6B3 A0A2J7PVM3 A0A1B6MJ99 A0A195D3V5 A0A2A4K4B3 A0A1B6GXZ2 A0A182FIS9 K7IU30 A0A224XGH6 Q16ZY5 A0A023EPJ1 E2C4N5 A0A0V0G980 F4W5L6 A0A0N7Z933 A0A158NSD4 A0A0L7QV45 A0A026WJP8 T1IBK9 A0A1B6D4X1 A0A161MZK5 A0A182GVC9 A0A023F371 A0A0P6J5T2 A0A023EPC0 B4NEJ5 A0A023EQF5 A0A146MDQ4 A0A182MN21 A0A146KSE4 A0A0K8T673 W8CBT6 F5HJM5 A0A2C9GPL9 A0A0A9XL74 Q9VKB8 A0A1W4VAZ0 B4P1Y1 A0A023ERM3 B3N3X7 A0A0J9R152 A0A336LVA3 B0WZ24 A0A0C9PL38 A0A034WN98 Q5BID6 A0A1Q3FII8 A0A182TBP2 E0W476 A0A0A1WXB0 A0A034WKG7 B4L823 A0A0K8VRM3 A0A2M4CXL7 A0A1I8PPZ8 D3TRJ0 B4N7A2 B3MVI2 A0A2M4BUP4 A0A2M4CXM9 A0A0K8UQP6 A0A2M3Z8X6 A0A0K8UNJ4 A0A1B0EX60 A0A1B0BV62 A0A1A9XFP3
A0A212EHH8 A0A2A4K3Z4 A0A084VNW2 A0A0N1IP16 A0A2C9GPE1 Q7PRW2 A0A2H1VJQ9 A0A1Q3G5K6 A0A1B6C109 W5JLV1 B0WZ23 A0A1Q3FQL1 A0A182PSU8 A0A182RHK9 A0A182L878 A0A182QFY3 A0A182MHI6 A0A182TE30 A0A182YH69 A0A067QTW2 A0A182N8G2 A0A182W5M3 A0A182XBY7 A0A182UQG1 A0A1L8DKW9 A0A182IZ06 A0A1B6H6B3 A0A2J7PVM3 A0A1B6MJ99 A0A195D3V5 A0A2A4K4B3 A0A1B6GXZ2 A0A182FIS9 K7IU30 A0A224XGH6 Q16ZY5 A0A023EPJ1 E2C4N5 A0A0V0G980 F4W5L6 A0A0N7Z933 A0A158NSD4 A0A0L7QV45 A0A026WJP8 T1IBK9 A0A1B6D4X1 A0A161MZK5 A0A182GVC9 A0A023F371 A0A0P6J5T2 A0A023EPC0 B4NEJ5 A0A023EQF5 A0A146MDQ4 A0A182MN21 A0A146KSE4 A0A0K8T673 W8CBT6 F5HJM5 A0A2C9GPL9 A0A0A9XL74 Q9VKB8 A0A1W4VAZ0 B4P1Y1 A0A023ERM3 B3N3X7 A0A0J9R152 A0A336LVA3 B0WZ24 A0A0C9PL38 A0A034WN98 Q5BID6 A0A1Q3FII8 A0A182TBP2 E0W476 A0A0A1WXB0 A0A034WKG7 B4L823 A0A0K8VRM3 A0A2M4CXL7 A0A1I8PPZ8 D3TRJ0 B4N7A2 B3MVI2 A0A2M4BUP4 A0A2M4CXM9 A0A0K8UQP6 A0A2M3Z8X6 A0A0K8UNJ4 A0A1B0EX60 A0A1B0BV62 A0A1A9XFP3
PDB
1YB1
E-value=5.93399e-49,
Score=489
Ontologies
KEGG
GO
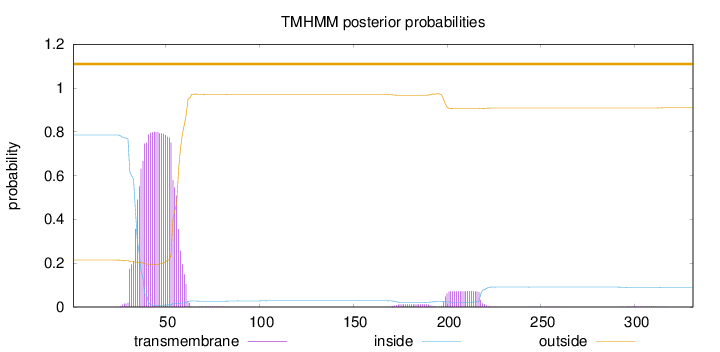
Topology
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
19.58173
Exp number, first 60 AAs:
17.66868
Total prob of N-in:
0.78543
POSSIBLE N-term signal
sequence
outside
1 - 331
Population Genetic Test Statistics
Pi
22.389987
Theta
23.206224
Tajima's D
-1.137943
CLR
1.331531
CSRT
0.111694415279236
Interpretation
Uncertain
