Gene
KWMTBOMO03092 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005812
Annotation
PREDICTED:_arginine_kinase_isoform_X2_[Bombyx_mori]
Full name
Arginine kinase
Location in the cell
Cytoplasmic Reliability : 2.546
Sequence
CDS
ATGGACGAAAATTTTCACCGAGCCGCTGTAGCGACTGTGGTCTCCCGTAATAGTGACAGCGGGACTGGATCTACTGTAGTTGACATAGCCAAAATCGCTGAATCCGTACTGGCGCGAGATCTGCCATCCAGGCTGGCATCGGGAGGGAAGGCCAAGGTTGAACGCGAGGGTGGCGGGGCGCGGGACGATGACGCGGGCGGGCTTAACGTACTGCGCGCTCCCGCACCAACCATTCTGCACCTAGCAGTCGGTTCTGACGGTTGTTCAAGTGCCAGAAAAGCCGCAACAATGGTCGACGCCGCAACCCTCGAGAAATTGGAGGCTGGTTTCAGCAAGCTCCAGGGATCCGACTCTAAGTCGCTGCTGAAGAAGTACCTTACCAGGGAAGTATTCGACAGCCTGAAGAACAAAAAGACCTCATTCGGATCCACCCTCCTTGACTGCATCCAATCGGGTGTCGAGAACTTGGACTCCGGCGTCGGTATCTACGCGCCGGACGCCGAGTCGTACTCCGTGTTCGCCGAGCTGTTTGACCCGATCATCGAGGACTACCACAATGGCTTCAAGAAGACCGACAAGCACCCGCCCAAGAACTGGGGAGACGTGGACACGCTCGGCAACCTCGACCCCGCCGGCGAGTTCGTGGTCTCCACCCGCGTGCGCTGCGGCCGCTCGCTCGAGGGGTACCCCTTCAACCCCTGCCTCACCGAGTCCCAGTACAAGGAGATGGAGGACAAGGTCTCCGGCACCCTGTCCAGCCTCGAGGGCGAGCTCAAGGGCACGTTCTACCCCCTCACCGGCATGTCGAAGGAGACCCAGCAGCAGCTCATCGACGACCACTTCCTGTTCAAGGAGGGCGACCGCTTCCTGCAGGCCGCCAACGCCTGCCGCTTCTGGCCCACCGGCCGCGGCATCTACCACAACGAGAACAAGACGTTCCTCGTGTGGTGCAACGAGGAGGACCACCTCCGCATCATCTCGATGCAGATGGGTGGCGACCTGCAGCAGGTATACAAGAGGCTGGTGAGCGCCGTCAACGAGATCGAGAAGAAGATCCCGTTCTCGCACCACGACCGGCTCGGCTTCCTCACGTTCTGCCCGACCAACCTGGGCACCACGGTCCGCGCCTCCGTGCACATCAAGCTGCCGAAGCTGGCGGCCGACAAGAAGAAGCTGGAGGAGGTGGCGTCCAAGTACCACCTGCAGGTCCGCGGCACGCGCGGCGAGCACACGGAGGCCGAGGGCGGCGTGTACGACATCTCCAACAAGCGCCGCATGGGGCTCACCGAGTACGACGCCGTCAAGGAGATGTACGACGGCATCGCCGAGCTCATCAAGATCGAGAAGTCTCTGTAG
Protein
MDENFHRAAVATVVSRNSDSGTGSTVVDIAKIAESVLARDLPSRLASGGKAKVEREGGGARDDDAGGLNVLRAPAPTILHLAVGSDGCSSARKAATMVDAATLEKLEAGFSKLQGSDSKSLLKKYLTREVFDSLKNKKTSFGSTLLDCIQSGVENLDSGVGIYAPDAESYSVFAELFDPIIEDYHNGFKKTDKHPPKNWGDVDTLGNLDPAGEFVVSTRVRCGRSLEGYPFNPCLTESQYKEMEDKVSGTLSSLEGELKGTFYPLTGMSKETQQQLIDDHFLFKEGDRFLQAANACRFWPTGRGIYHNENKTFLVWCNEEDHLRIISMQMGGDLQQVYKRLVSAVNEIEKKIPFSHHDRLGFLTFCPTNLGTTVRASVHIKLPKLAADKKKLEEVASKYHLQVRGTRGEHTEAEGGVYDISNKRRMGLTEYDAVKEMYDGIAELIKIEKSL
Summary
Catalytic Activity
ATP + L-arginine = ADP + H(+) + N(omega)-phospho-L-arginine
Similarity
Belongs to the ATP:guanido phosphotransferase family.
Keywords
Allergen
ATP-binding
Kinase
Nucleotide-binding
Transferase
Feature
chain Arginine kinase
Uniprot
H9J8H0
K4PWX1
Q2F5T5
A9Z0V0
A0A194PP98
I4DIQ0
+ More
A0A0N1I6C4 A0A0L7LS98 I4DMK8 A0A2H1X344 A0A2A4JHS8 A0A1E1WQH9 S4PWZ5 A0A1E1WK34 E9NZS5 E3W086 I1SSI2 D3Y4D1 M4PZR4 D5KZY5 A0A3B0ENM0 A0A1E1W127 B1NLE2 C8CBB4 A0A212EKU4 A0A0U3MZT4 D9YT56 F4YCW1 Q95PM9 A0A1E1W0W2 A0A0A0YWU7 A0A0A7DWF3 A0A0P0M031 A0A1W4WYG0 A0A1W4WNQ5 A0A0L7QU24 A0A1W4WZH4 A0A1Q3FM56 A0A182GZB5 A0A1W4WZQ9 A0A1Q3EUQ0 A0A1B0GH56 A0A182IY03 A0A1L8E0Y1 A0A2J7R607 A0A232F9M6 A0A023ERL8 A0A1Q3FMT5 A0A0N0BJZ8 A0A1S4GQH2 A0A182QGC9 A0A1Q3FMR9 A0A0T6B5L1 G3FP78 B0WLS0 A0A084W9Z4 R4G5I9 A0A1B1MRP7 K7IUI5 A0A182FDN2 Q1HR67 A0A182GN72 A0A1D1XCV8 V9IDW3 A0A1Q3FMQ9 A1KY39 B9VAT1 A0A2A3EGD2 C7S2Z9 C6H0K8 A0A1L8E0F8 E1U7W2 A0A182UUG9 A0A3F2YWB7 A0A139WNX9 A0A182NGU6 A0A182HWU3 A0A182XMY7 A0A0C9Q8D2 A0A182U5Z3 A0A2M4A3B1 A0A182MLL4 A0A182JV73 A5HUI6 A0A067RAL9 D3JUE7 A0A182LJ63 A0A2M3YYL8 A0A069DSB2 T1E1J2 F6GPJ8 T1DSI6 A0A336LGY4 Q7PIQ5 G8XWV9 A0A2R7WRQ5 N6U6N6 A0A2M3YYN8 A0A291L1R7 J3JXG0
A0A0N1I6C4 A0A0L7LS98 I4DMK8 A0A2H1X344 A0A2A4JHS8 A0A1E1WQH9 S4PWZ5 A0A1E1WK34 E9NZS5 E3W086 I1SSI2 D3Y4D1 M4PZR4 D5KZY5 A0A3B0ENM0 A0A1E1W127 B1NLE2 C8CBB4 A0A212EKU4 A0A0U3MZT4 D9YT56 F4YCW1 Q95PM9 A0A1E1W0W2 A0A0A0YWU7 A0A0A7DWF3 A0A0P0M031 A0A1W4WYG0 A0A1W4WNQ5 A0A0L7QU24 A0A1W4WZH4 A0A1Q3FM56 A0A182GZB5 A0A1W4WZQ9 A0A1Q3EUQ0 A0A1B0GH56 A0A182IY03 A0A1L8E0Y1 A0A2J7R607 A0A232F9M6 A0A023ERL8 A0A1Q3FMT5 A0A0N0BJZ8 A0A1S4GQH2 A0A182QGC9 A0A1Q3FMR9 A0A0T6B5L1 G3FP78 B0WLS0 A0A084W9Z4 R4G5I9 A0A1B1MRP7 K7IUI5 A0A182FDN2 Q1HR67 A0A182GN72 A0A1D1XCV8 V9IDW3 A0A1Q3FMQ9 A1KY39 B9VAT1 A0A2A3EGD2 C7S2Z9 C6H0K8 A0A1L8E0F8 E1U7W2 A0A182UUG9 A0A3F2YWB7 A0A139WNX9 A0A182NGU6 A0A182HWU3 A0A182XMY7 A0A0C9Q8D2 A0A182U5Z3 A0A2M4A3B1 A0A182MLL4 A0A182JV73 A5HUI6 A0A067RAL9 D3JUE7 A0A182LJ63 A0A2M3YYL8 A0A069DSB2 T1E1J2 F6GPJ8 T1DSI6 A0A336LGY4 Q7PIQ5 G8XWV9 A0A2R7WRQ5 N6U6N6 A0A2M3YYN8 A0A291L1R7 J3JXG0
EC Number
2.7.3.3
Pubmed
EMBL
BABH01027400
BABH01027401
AB711181
BAM62792.1
DQ272299
DQ311338
+ More
EU350575 FJ013046 AB711180 ABB88514.1 ABD36282.1 ABY59719.1 ACI01048.1 BAM62791.1 EU327675 ABY28386.1 KQ459597 KPI95132.1 AK401168 BAM17790.1 KQ460847 KPJ11986.1 JTDY01000196 KOB78348.1 AK402526 BAM19148.1 ODYU01013088 SOQ59770.1 NWSH01001344 PCG71607.1 GDQN01001973 JAT89081.1 GAIX01005928 JAA86632.1 GDQN01003833 JAT87221.1 HQ840714 ADW94627.1 HQ336337 ADO23657.1 HM068068 AEB71991.1 GU396008 JN185455 ADD22718.1 AET41718.1 KC262639 KC262642 AGH14259.1 AGH14262.1 GU726905 ADE27964.1 PCG71608.1 RBVL01000226 RKO10391.1 GDQN01010354 GDQN01003477 GDQN01003447 JAT80700.1 JAT87577.1 JAT87607.1 EF600057 ABU98622.1 GQ379235 ACU68932.1 AGBW02014213 OWR42095.1 KU201285 ALX37956.1 GU060292 HQ327310 ADC29631.1 ADO32583.1 GU937510 ADW23117.1 AJ315030 GDQN01010429 JAT80625.1 KJ728856 AIX48027.1 KJ019020 AIY31784.1 KR869785 ALK82255.1 KQ414736 KOC62125.1 GFDL01006384 JAV28661.1 JXUM01002968 JXUM01002969 JXUM01002970 KQ560148 KXJ84163.1 GFDL01016008 JAV19037.1 AJWK01003984 GFDF01001745 JAV12339.1 NEVH01006986 PNF36266.1 NNAY01000680 OXU27017.1 GAPW01002269 JAC11329.1 GFDL01006229 JAV28816.1 KQ435710 KOX79881.1 AAAB01008960 AXCN02000366 GFDL01006227 JAV28818.1 LJIG01009680 KRT82596.1 JF751027 AEO51760.1 DS231990 EDS30648.1 ATLV01021973 KE525326 KFB47038.1 ACPB03003141 GAHY01000520 JAA76990.1 KU521364 ANS71244.1 AAZX01005759 AAZX01013667 DQ440227 CH477562 ABF18260.1 EAT38978.1 JXUM01075994 KQ562917 KXJ74851.1 GDJX01027672 JAT40264.1 JR039057 JR039059 AEY58509.1 AEY58510.1 GFDL01006233 JAV28812.1 AY563004 AAT77152.1 FJ514482 ACM24358.1 KZ288266 PBC30216.1 DQ886906 ABJ88949.1 FN421429 FN421431 CAZ65719.1 CAZ65721.1 GFDF01001903 JAV12181.1 FJ882065 ACZ68114.1 KY971526 KQ971307 AVD69630.1 KYB29642.1 APCN01001538 GBYB01010493 JAG80260.1 GGFK01001807 MBW35128.1 AXCM01000869 AB264171 BAF62631.1 KK852580 KDR20892.1 GU301882 ADB92491.1 GGFM01000622 MBW21373.1 GBGD01001941 JAC86948.1 GALA01001619 JAA93233.1 EU702488 ACH61778.1 GAMD01001464 JAB00127.1 UFQT01000004 SSX17244.1 EAA44056.3 EDO63704.1 FJ855501 AEV23883.1 KK855169 PTY21245.1 APGK01047206 APGK01047207 KB741077 KB631792 ENN74227.1 ERL86145.1 GGFM01000623 MBW21374.1 KY809862 ATI09810.1 BT127929 AEE62891.1
EU350575 FJ013046 AB711180 ABB88514.1 ABD36282.1 ABY59719.1 ACI01048.1 BAM62791.1 EU327675 ABY28386.1 KQ459597 KPI95132.1 AK401168 BAM17790.1 KQ460847 KPJ11986.1 JTDY01000196 KOB78348.1 AK402526 BAM19148.1 ODYU01013088 SOQ59770.1 NWSH01001344 PCG71607.1 GDQN01001973 JAT89081.1 GAIX01005928 JAA86632.1 GDQN01003833 JAT87221.1 HQ840714 ADW94627.1 HQ336337 ADO23657.1 HM068068 AEB71991.1 GU396008 JN185455 ADD22718.1 AET41718.1 KC262639 KC262642 AGH14259.1 AGH14262.1 GU726905 ADE27964.1 PCG71608.1 RBVL01000226 RKO10391.1 GDQN01010354 GDQN01003477 GDQN01003447 JAT80700.1 JAT87577.1 JAT87607.1 EF600057 ABU98622.1 GQ379235 ACU68932.1 AGBW02014213 OWR42095.1 KU201285 ALX37956.1 GU060292 HQ327310 ADC29631.1 ADO32583.1 GU937510 ADW23117.1 AJ315030 GDQN01010429 JAT80625.1 KJ728856 AIX48027.1 KJ019020 AIY31784.1 KR869785 ALK82255.1 KQ414736 KOC62125.1 GFDL01006384 JAV28661.1 JXUM01002968 JXUM01002969 JXUM01002970 KQ560148 KXJ84163.1 GFDL01016008 JAV19037.1 AJWK01003984 GFDF01001745 JAV12339.1 NEVH01006986 PNF36266.1 NNAY01000680 OXU27017.1 GAPW01002269 JAC11329.1 GFDL01006229 JAV28816.1 KQ435710 KOX79881.1 AAAB01008960 AXCN02000366 GFDL01006227 JAV28818.1 LJIG01009680 KRT82596.1 JF751027 AEO51760.1 DS231990 EDS30648.1 ATLV01021973 KE525326 KFB47038.1 ACPB03003141 GAHY01000520 JAA76990.1 KU521364 ANS71244.1 AAZX01005759 AAZX01013667 DQ440227 CH477562 ABF18260.1 EAT38978.1 JXUM01075994 KQ562917 KXJ74851.1 GDJX01027672 JAT40264.1 JR039057 JR039059 AEY58509.1 AEY58510.1 GFDL01006233 JAV28812.1 AY563004 AAT77152.1 FJ514482 ACM24358.1 KZ288266 PBC30216.1 DQ886906 ABJ88949.1 FN421429 FN421431 CAZ65719.1 CAZ65721.1 GFDF01001903 JAV12181.1 FJ882065 ACZ68114.1 KY971526 KQ971307 AVD69630.1 KYB29642.1 APCN01001538 GBYB01010493 JAG80260.1 GGFK01001807 MBW35128.1 AXCM01000869 AB264171 BAF62631.1 KK852580 KDR20892.1 GU301882 ADB92491.1 GGFM01000622 MBW21373.1 GBGD01001941 JAC86948.1 GALA01001619 JAA93233.1 EU702488 ACH61778.1 GAMD01001464 JAB00127.1 UFQT01000004 SSX17244.1 EAA44056.3 EDO63704.1 FJ855501 AEV23883.1 KK855169 PTY21245.1 APGK01047206 APGK01047207 KB741077 KB631792 ENN74227.1 ERL86145.1 GGFM01000623 MBW21374.1 KY809862 ATI09810.1 BT127929 AEE62891.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000218220
UP000276569
+ More
UP000007151 UP000192223 UP000053825 UP000069940 UP000249989 UP000092461 UP000075880 UP000235965 UP000215335 UP000053105 UP000075886 UP000002320 UP000030765 UP000015103 UP000002358 UP000069272 UP000008820 UP000242457 UP000075903 UP000075885 UP000007266 UP000075884 UP000075840 UP000076407 UP000075902 UP000075883 UP000075881 UP000027135 UP000075882 UP000007062 UP000019118 UP000030742
UP000007151 UP000192223 UP000053825 UP000069940 UP000249989 UP000092461 UP000075880 UP000235965 UP000215335 UP000053105 UP000075886 UP000002320 UP000030765 UP000015103 UP000002358 UP000069272 UP000008820 UP000242457 UP000075903 UP000075885 UP000007266 UP000075884 UP000075840 UP000076407 UP000075902 UP000075883 UP000075881 UP000027135 UP000075882 UP000007062 UP000019118 UP000030742
Interpro
Gene 3D
ProteinModelPortal
H9J8H0
K4PWX1
Q2F5T5
A9Z0V0
A0A194PP98
I4DIQ0
+ More
A0A0N1I6C4 A0A0L7LS98 I4DMK8 A0A2H1X344 A0A2A4JHS8 A0A1E1WQH9 S4PWZ5 A0A1E1WK34 E9NZS5 E3W086 I1SSI2 D3Y4D1 M4PZR4 D5KZY5 A0A3B0ENM0 A0A1E1W127 B1NLE2 C8CBB4 A0A212EKU4 A0A0U3MZT4 D9YT56 F4YCW1 Q95PM9 A0A1E1W0W2 A0A0A0YWU7 A0A0A7DWF3 A0A0P0M031 A0A1W4WYG0 A0A1W4WNQ5 A0A0L7QU24 A0A1W4WZH4 A0A1Q3FM56 A0A182GZB5 A0A1W4WZQ9 A0A1Q3EUQ0 A0A1B0GH56 A0A182IY03 A0A1L8E0Y1 A0A2J7R607 A0A232F9M6 A0A023ERL8 A0A1Q3FMT5 A0A0N0BJZ8 A0A1S4GQH2 A0A182QGC9 A0A1Q3FMR9 A0A0T6B5L1 G3FP78 B0WLS0 A0A084W9Z4 R4G5I9 A0A1B1MRP7 K7IUI5 A0A182FDN2 Q1HR67 A0A182GN72 A0A1D1XCV8 V9IDW3 A0A1Q3FMQ9 A1KY39 B9VAT1 A0A2A3EGD2 C7S2Z9 C6H0K8 A0A1L8E0F8 E1U7W2 A0A182UUG9 A0A3F2YWB7 A0A139WNX9 A0A182NGU6 A0A182HWU3 A0A182XMY7 A0A0C9Q8D2 A0A182U5Z3 A0A2M4A3B1 A0A182MLL4 A0A182JV73 A5HUI6 A0A067RAL9 D3JUE7 A0A182LJ63 A0A2M3YYL8 A0A069DSB2 T1E1J2 F6GPJ8 T1DSI6 A0A336LGY4 Q7PIQ5 G8XWV9 A0A2R7WRQ5 N6U6N6 A0A2M3YYN8 A0A291L1R7 J3JXG0
A0A0N1I6C4 A0A0L7LS98 I4DMK8 A0A2H1X344 A0A2A4JHS8 A0A1E1WQH9 S4PWZ5 A0A1E1WK34 E9NZS5 E3W086 I1SSI2 D3Y4D1 M4PZR4 D5KZY5 A0A3B0ENM0 A0A1E1W127 B1NLE2 C8CBB4 A0A212EKU4 A0A0U3MZT4 D9YT56 F4YCW1 Q95PM9 A0A1E1W0W2 A0A0A0YWU7 A0A0A7DWF3 A0A0P0M031 A0A1W4WYG0 A0A1W4WNQ5 A0A0L7QU24 A0A1W4WZH4 A0A1Q3FM56 A0A182GZB5 A0A1W4WZQ9 A0A1Q3EUQ0 A0A1B0GH56 A0A182IY03 A0A1L8E0Y1 A0A2J7R607 A0A232F9M6 A0A023ERL8 A0A1Q3FMT5 A0A0N0BJZ8 A0A1S4GQH2 A0A182QGC9 A0A1Q3FMR9 A0A0T6B5L1 G3FP78 B0WLS0 A0A084W9Z4 R4G5I9 A0A1B1MRP7 K7IUI5 A0A182FDN2 Q1HR67 A0A182GN72 A0A1D1XCV8 V9IDW3 A0A1Q3FMQ9 A1KY39 B9VAT1 A0A2A3EGD2 C7S2Z9 C6H0K8 A0A1L8E0F8 E1U7W2 A0A182UUG9 A0A3F2YWB7 A0A139WNX9 A0A182NGU6 A0A182HWU3 A0A182XMY7 A0A0C9Q8D2 A0A182U5Z3 A0A2M4A3B1 A0A182MLL4 A0A182JV73 A5HUI6 A0A067RAL9 D3JUE7 A0A182LJ63 A0A2M3YYL8 A0A069DSB2 T1E1J2 F6GPJ8 T1DSI6 A0A336LGY4 Q7PIQ5 G8XWV9 A0A2R7WRQ5 N6U6N6 A0A2M3YYN8 A0A291L1R7 J3JXG0
PDB
4BG4
E-value=6.38989e-173,
Score=1560
Ontologies
GO
PANTHER
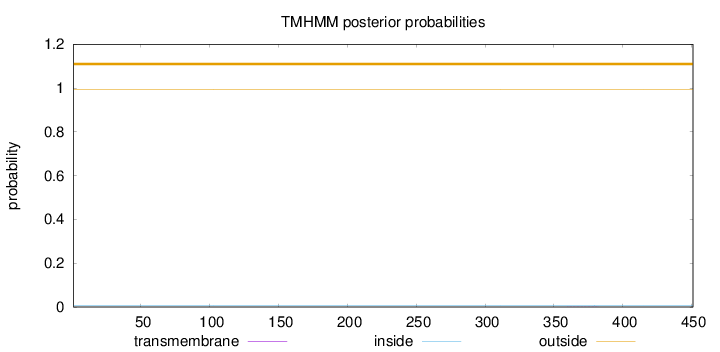
Topology
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0102
Exp number, first 60 AAs:
0.00055
Total prob of N-in:
0.00643
outside
1 - 451
Population Genetic Test Statistics
Pi
16.235508
Theta
20.144673
Tajima's D
-0.699042
CLR
2.752886
CSRT
0.201989900504975
Interpretation
Uncertain
